- Departments and Units
- Majors and Minors
- LSA Course Guide
- LSA Gateway

Search: {{$root.lsaSearchQuery.q}}, Page {{$root.page}}
| {{item.snippet}} |
- Accessibility
- Undergraduates
- Instructors
- Alums & Friends

- ★ Writing Support
- Minor in Writing
- First-Year Writing Requirement
- Transfer Students
- Writing Guides
- Peer Writing Consultant Program
- Upper-Level Writing Requirement
- Writing Prizes
- International Students
- ★ The Writing Workshop
- Dissertation ECoach
- Fellows Seminar
- Dissertation Writing Groups
- Rackham / Sweetland Workshops
- Dissertation Writing Institute
- Guides to Teaching Writing
- Teaching Support and Services
- Support for FYWR Courses
- Support for ULWR Courses
- Writing Prize Nominating
- Alums Gallery
- Commencement
- Giving Opportunities
- How Do I Present Findings From My Experiment in a Report?
- How Do I Make Sure I Understand an Assignment?
- How Do I Decide What I Should Argue?
- How Can I Create Stronger Analysis?
- How Do I Effectively Integrate Textual Evidence?
- How Do I Write a Great Title?
- What Exactly is an Abstract?
- What is a Run-on Sentence & How Do I Fix It?
- How Do I Check the Structure of My Argument?
- How Do I Write an Intro, Conclusion, & Body Paragraph?
- How Do I Incorporate Quotes?
- How Can I Create a More Successful Powerpoint?
- How Can I Create a Strong Thesis?
- How Can I Write More Descriptively?
- How Do I Incorporate a Counterargument?
- How Do I Check My Citations?
See the bottom of the main Writing Guides page for licensing information.
Many believe that a scientist’s most difficult job is not conducting an experiment but presenting the results in an effective and coherent way. Even when your methods and technique are sound and your notes are comprehensive, writing a report can be a challenge because organizing and communicating scientific findings requires patience and a thorough grasp of certain conventions. Having a clear understanding of the typical goals and strategies for writing an effective lab report can make the process much less troubling.
General Considerations
It is useful to note that effective scientific writing serves the same purpose that your lab report should. Good scientific writing explains:
- The goal(s) of your experiment
- How you performed the experiment
- The results you obtained
- Why these results are important
While it’s unlikely that you’re going to win the Nobel Prize for your work in an undergraduate laboratory course, tailoring your writing strategies in imitation of professional journals is easier than you might think, since they all follow a consistent pattern. However, your instructor has the final say in determining how your report should be structured and what should appear in each section. Please use the following explanations only to supplement your given writing criteria, rather than thinking of them as an indication of how all lab reports must be written.
Analysis and presentation of experimental results : with examples, problems and programs
Available online.
- SpringerLink
More options
- Find it at other libraries via WorldCat
- Contributors
Description
Creators/contributors, contents/summary.
- Basic statistical concepts.- Measurement errors.- A though experiment.- The statistical analysis of experimental results.- The presentation of numerical results.- The propagation of errors.- The three basic probability distributions.- The statistics of radioactivity.- Elements from the theory of errors.- Comparison and rejection of measurements.- The method of least squares.- Graphs.- The written report of the results of an experiment.- Appendix 1. Least squares straight line y = + x . The errors in and
- Appendix 2. Dimensional analysis.- Appendix 3. The use of random numbers in finding values of a variable x which are distributed according to a given probability density function f(x).- Appendix 4. Values of fundamental physical constants.- Answers to the problems.- List of programs and code samples.- Index.
- (source: Nielsen Book Data)
- Basic statistical concepts
- Measurement errors
- A though experiment
- The statistical analysis of experimental results
- The presentation of numerical results
- The propagation of errors
- The three basic probability distributions
- The statistics of radioactivity
- Elements from the theory of errors
- Comparison and rejection of measurements
- The method of least squares
- The written report of the results of an experiment
- Appendix 1. Least squares straight line y =α + λ x . The errors in α and λ
- Appendix 2. Dimensional analysis
- Appendix 3. The use of random numbers in finding values of a variable x which are distributed according to a given probability density function f(x)
- Appendix 4. Values of fundamental physical constants
- Answers to the problems
- List of programs and code samples
Bibliographic information
Browse related items.
- Stanford Home
- Maps & Directions
- Search Stanford
- Emergency Info
- Terms of Use
- Non-Discrimination
- Accessibility
© Stanford University , Stanford , California 94305 .

Journals, books & databases
- Author & reviewer hub
- Author guidelines & information
- Prepare your article
- Experimental reporting requirements

Experimental details and characterisation required for journal articles
Guidance on reporting experimental procedures and compound characterisation

We believe that where possible, all data associated with the research in a manuscript should be Findable, Accessible, Interoperable and Reusable (FAIR), enabling other researchers to replicate and build on that research. We strongly encourage authors to deposit the data underpinning their research in appropriate repositories and make it as openly accessible as possible.
For all submissions to our journals, any data required to understand and verify the research in an article must be made available on submission. To comply, we suggest authors deposit their data in an appropriate repository. Where this isn’t possible, we ask authors to include the data as part of the article Supplementary Information. If necessary data are not made available, authors may be requested to provide these as part of the peer-review process, or in light of any post-publication concerns.
Please see our Data sharing guidance and policy for more details on specific data types and recommended repositories.
Some journals may also have additional subject requirements for both sharing and/or publishing supporting data, so please ensure you check the journal-specific guidelines.
On this page
General guidance, presentation of experimental data, post-acquisition processing of data.
- Data citation
Human and animal welfare
Biomolecules, characterisation of compounds and materials, computational studies and modelling, electrophoretic gels and blots, fluorescence sensors, inorganic and organometallic compounds, macromolecular structure and sequence data.
- Magnetic measurements
Nanomaterials
Organic compounds, polymers and macromolecules, synthetic procedures, system models, x-ray crystallography, view all guidelines for preparing and formatting your article.
Please note, these guidelines are relevant to all of our journals. Make sure that you check your chosen journal’s web pages for specific guidelines too.
These experimental reporting requirements apply to both new compounds and known compounds prepared by a new or modified method.
It is the authors’ responsibility to provide descriptions of the experiments in enough detail to enable other skilled researchers to accurately reproduce the work.
Experimental procedures, compound characterization data, research materials necessary to enable the reproduction of an experiment and references to the associated literature should be provided in the experimental section of the manuscript.
Standard techniques and methods used throughout the work should be stated at the beginning of the experimental section; descriptions of these are not needed.
For known compounds synthesised via a literature procedure, authors should provide a reference to previously published characterization data.
Sources of starting materials obtained need not be identified unless the compound is not widely available, or the source is critical for the experimental result. Only non-standard apparatus should be described and commercially available instruments can be referred to by their stock numbers.
The accuracy of primary measurements should be stated. Figures should include error bars where appropriate, and results should be accompanied by an analysis of experimental uncertainty. Care should be taken to report the correct number of significant figures throughout the manuscript.
Any unusual hazards associated with the chemicals, procedures or equipment should be clearly identified.
For studies that involve the use of live animals or human subjects please refer to our Human and Animal Welfare policy .
Please see the sections below for detailed information about how to present specific types of data.
Data associated with particular compounds should be listed after the name of the compound concerned, following the description of its preparation. If comparison is to be made with literature values, these should be quoted in parentheses - for example, mp 157 °C (from chloroform) (lit., 19 156 °C), or ν max /cm -1 2020 and 1592 (lit., 24 2015 and 1600).
The suggested order in which the most commonly encountered data for a new compound should be cited follows:
Melting point
Optical rotation, refractive index, elemental analysis, uv absorptions, ir absorptions.
- NMR spectrum
- Mass spectrum
You can find more information about each of these below:
The following information is a guide to the presentation of experimental data, including appropriate formats for citation.
Yield should be presented in parentheses after the compound name (or its equivalent). Weight and percentage should be separated by a comma – for example, the lactone (7.1 g, 56%).
The melting point should be presented in the form mp 75 °C (from EtOH) - that is, the crystallisation solvent in parentheses. If an identical mixed melting point is to be recorded, the form mp and mixed mp 75 °C is appropriate.
The units should be stated in the preamble to the Experimental section – for example, [ α ] D values are given in 10 −1 deg cm 2 g −1 . This should be shown in the form [α] D 22–22.5 (c 0.95 in EtOH) – that is, concentration and solvent in parentheses.
Given in the form n D 22 1.653.
For the presentation of elemental analyses, both forms (Found: C, 63.1; H, 5.4. C 13 H 13 NO 4 requires C, 63.2; H, 5.3%) and (Found: C, 62.95; H, 5.4. Calc. for C 13 H 13 NO 4 : C, 63.2; H, 5.3%) are acceptable. Analyses are normally quoted to the nearest 0.1%, but a 5 in the second place of decimals is retained.
If a molecular weight is to be included, the appropriate form is: [Found: C, 63.1; H, 5.4%; M (mass spectrum), 352 (or simply M+, 352). C 13 H 13 NO 4 requires C, 63.2; H, 5.3%; M, 352].
We encourage authors to provide instrumental details and the chromatograms of the performed measurements in the Supplementary Information where possible.
These should be given in the form λmax(EtOH)/nm 228 (ε/dm 3 mol -1 cm -1 40 900), 262 (19 200) and 302 (11 500). Inflections and shoulders are specified as 228infl or 262sh. Alternatively the following form may be used: λmax (EtOH)/nm 228, 262 and 302 (ε/dm 3 mol -1 cm -1 40 900, 19 200 and 11 500); log ε may be quoted instead of ε.
IR absorption should be presented as follows: ν max /cm -1 3460 and 3330 (NH), 2200 (conj. CN), 1650 (CO) and 1620 (CN). The type of signal (s, w, vs, br) can be indicated by appended letters (for example 1760vs).
For all NMR spectra δ values should be used, with the nucleus indicated by subscript if necessary (for example, δ H , δ C ). A statement specifying the units of the coupling constants should be given in the preamble to the Experimental section – for example, J values are given in Hz. Instrument frequency, solvent, and standard should be specified. For example: δ H (100 MHz; CDCl3; Me4Si) 2.3 (3 H, s, Me), 2.5 (3 H, s, COMe), 3.16 (3 H, s, NMe) and 7.3–7.6 (5 H, m, Ph).
A broad signal may be denoted by br, such as 2.43 (1 H, br s, NH). Order of citation in parentheses: (i) number of equivalent nuclei (by integration), (ii) multiplicity (s, d, t, q), (iii) coupling constant – for example, J 1,2 2, J A B 4, (iv) assignment; italicisation can be used to specify the nuclei concerned (for example, CH3CH2). The proton attached to C-6 may be designated C(6)H or 6-H; the methyl attached to C-6, 6-Me or C(6)Me.
Mutually coupled protons in 1 H NMR spectra should be quoted with precisely matching J values, in order to assist thorough interpretation. In instances of any ambiguities when taking readings from computer printouts, mean J values should be quoted, rounded to the nearest decimal point.
Mass spectrometry data
Mass spectrometry data should be given in the form: m/z 183 (M + , 41%), 168 (38), 154 (9), 138 (31) etc. The molecular ion may be specified as shown if desired. Relative intensities should be shown in parentheses (% only included once). Other assignments may be included in the form m/z 152 (33, M − CH 3 CONH 2 ). Metastable peaks may be listed as: M* 160 (189→174), 147 (176→161), etc. The type of spectrum (field desorption, electron impact, etc.) should be indicated. Exact masses quoted for identification purposes should be accurate to within 5 ppm (EI and CI) or 10 ppm (FAB or LSIMS).
Authors might be asked during peer review to provide the original unprocessed data to the editors/reviewers of the journal.
All image acquisition and processing tools (including their settings) should be clearly stated in the manuscript. The amount of post-acquisition processing of data should be kept to a minimum. Any type of alteration such as image processing, cropping and groupings should be clearly stated in the figure caption and the Supplementary Information (SI) - clearly describing the process of alteration. Data manipulation (for example, normalisation or handling of missing values) should be noted.
Image processing changes should be applied to the entire image as well as all other images it is compared to. Processed images should still represent all the original data (with no data missing) and touch-up tools should be avoided.
Genuine and relevant signals in spectra should not be lost due to image enhancement.
Microscopy images of cells from multiple fields should not be compared but shown as single images (at least as part of the deposited data or in the SI).
Data Citation
For author-generated datasets that are directly associated with the article, we encourage authors to add data citations as bibliographic references within the article and the Data Availability Statement (DAS). Within the DAS, the citation should be given alongside information on datasets associated with the study and where to find them.
For datasets associated with previous studies, we encourage authors to add data citations as bibliographic references within the main text as they are mentioned. Data citation is encouraged as an alternative to informal references or mentions of local identifiers.
Suggested reference format for data citations:
[A. Name, B. Name and C. Name], [Name of repository / type of dataset], [Deposition number], [Year], [DOI, or URL if not available, of the dataset].
An example:
P. Cui, D. P. McMahon, P. R. Spackman, B. M. Alston, M. A. Little, G. M. Day and A. I. Cooper, 2019, CCDC Experimental Crystal Structure Determination: 1915306, DOI: 10.5517/ccdc.csd.cc22912j
Please also refer to the guidelines from the relevant repository on which information to provide in a citation.
When a study involves the use of live animal subjects, authors should adhere to the Animal Research: Reporting In Vivo Experiments (ARRIVE) 2.0 guidelines. When a study involves the use of human subjects, authors should adhere to the general principles set out in the Declaration of Helsinki .
Authors must include in the "methods/experimental" section of the manuscript a statement that all experiments were performed in compliance with the relevant guidelines. The statement must name the institutional/local ethics committee that has approved the study, and where possible the approval or case number should be provided.
Details of all guidelines followed should be provided. A statement regarding informed consent is required for all studies involving human subjects. Reviewers may be asked to comment specifically on any cases in which concerns arise.
For studies involving the use of animal subjects, authors are encouraged to make the completed ARRIVE 2.0 checklist available during peer review, for example by sharing it as part of the Supplementary Information (SI) or citing the deposited item.
The journals’ editorial teams reserve the right to request additional information in relation to experiments on vertebrates or higher invertebrates as necessary for the evaluation of the manuscript e.g., in the context of appropriate animal welfare or studies that involve death as an experimental endpoint.
Authors and referees should note the following guidelines for articles reporting electrochemical data and setup of batteries. It is the authors’ responsibility to ensure that the following information is provided in the main manuscript or Supplementary Information as appropriate.
The setup used for electrochemical testing should be clearly specified in the Experimental Information. For example, full or half cells, reference electrode (if used), testing temperature, etc.
When reporting electrochemical performance data, the authors should clearly state how many experimental runs these data are based on. The electrochemical performance value calculations should be clearly explained (including information on using charging or discharging values). All electrochemical data should be reported to an appropriate number of significant figures, along with standard deviation and error bars on graphs.
When reporting electrode performance values , the thickness of the electrode and the mass percentage of all electrode components (active material, additive, binder, etc.), the total mass of the electrode, and the geometric area of the electrode should be provided.
When reporting device-level performance values, the mass percentage of all battery components (active material, additive, binder, casing, current collector, electrolyte, separator, etc.), the total mass of the battery, and the geometric area of the electrode should be reported.
The mass percent and theoretical capacity of the active material should be provided if the theoretical capacity of the studied material is known. The theoretical capacity should be used to calculate C-rate. Alternatively, a rigorous use of A g -1 is recommended.
Pre-cycling and/or first cycle data should be reported.
Calculations of battery capacity should report the capacity obtained (in mAh g -1 ; if appropriate, volumetric values can be added in the unit of mAh cm -3 ) with the cycling rate and at what cycle number this capacity was obtained clearly stated. Average capacities for ≥3 cells with standard deviation are preferred.
It is the authors’ responsibility to provide rigorous evidence for the identity and purity of the biomolecules (for example, enzymes, proteins, DNA/RNA, oligosaccharides, oligonucleotides) described.
The identity of the biomolecule should be substantiated by employing at least one appropriate method, which may include one or more of the following:
Mass spectrometry
- Sequencing data (for proteins and oligonucleotides)
- High field 1 H or 13 C NMR
The purity should be established by one or more of the following
- Gel electrophoresis
- Capillary electrophoresis
Sequence verification should also be carried out for nucleic acids in molecular biology, including all mutants; for new protein or gene sequences, the entire sequence should be provided. For organic synthesis involving DNA, RNA oligonucleotides, their derivatives or mimics, purity should be established using HPLC and mass spectrometry as a minimum.
Provide usual organic chemistry analytical requirements for the novel monomer ( see Organic compounds ). However, it is not necessary to provide this level of characterisation for the oligonucleotide into which the novel monomer is incorporated.
Provide sufficient detail to identify the species being used. Specific information on antibodies is essential. Commercial sources and, if new antibodies are generated, full experimental details such as immunogen/phage, species, protocols for mAb-) should be given. We strongly recommend authors use unique Resource Identifiers for model organisms, antibodies, and tools, and publish them with full descriptions.
Present scatter plots of data, sensitivity, and specificity values with confidence intervals and results of receiver operating characteristic curve analysis. If a marker is already routinely used for that disease, comparison with that marker should be included.
Where the screening of new catalysts is reported, authors should provide a mass balance for all reactions (using, for example, an internal standard in their analysis technique). Recycling efficiencies should be based on reaction rates measurements and not product yield as a function of cycle. It is highly desirable to report the reaction rate for the catalysts as turnover frequency or mass-specific activity or, for heterogeneous catalysts, as surface-specific activity.
It is the responsibility of authors to provide fully convincing evidence for the homogeneity and identity of all compounds whose preparations they describe. Evidence of both purity and identity is required to establish that the properties and constants reported are those of a compound as claimed.
Reviewers will assess the evidence in support of the homogeneity and structure of all new compounds. No hard and fast rules can be laid down to cover all types of compound, but evidence for the unequivocal identification of new compounds should, wherever possible, include good elemental analytical data – an accurate mass measurement of a molecular ion does not provide evidence of purity of a compound and should be accompanied by independent evidence of homogeneity.
Where elemental analytical data cannot be obtained, appropriate evidence that is convincing to an expert in the field may be acceptable. Normally, for diamagnetic compounds this entails, at a minimum, a high resolution mass spectrometry measurement along with assigned 1 H and/or 13 C NMR spectra devoid of visible impurities.
Spectroscopic information necessary for the assignment of structure should be given. How complete this information should be depends upon the circumstances; the structure of a compound obtained from an unusual reaction or isolated from a natural source should be supported by stronger evidence than one that was produced by a standard reaction from a precursor of undisputed structure.
Particular care should be taken in supporting the assignments of stereochemistry (both relative and absolute) of chiral compounds reported, for example by one of the following:
- NMR spectroscopy
- Polarimetry
- Correlation with known compounds of undisputed configuration
In cases where mixtures of isomers are generated (for example, E-Z isomers, enantiomers, diastereoisomers), the constitution of the mixture should usually be established using appropriate analytical techniques (for example, NMR spectroscopy, GC, HPLC) and reported in an unambiguous fashion.
For an asymmetric reaction in which an enantiomeric mixture is prepared, the direct measurement of the enantiomer ratio expressed as the enantiomeric excess (ee) is recommended, and is preferred to less reliable polarimetry methods.
If a compound is new more detailed characterisation will be required. A compound is considered to be new if:
- it has not been prepared before
- it has been prepared before but not adequately purified
- it has been purified but not adequately characterised
- it has been assigned an erroneous composition previously
- it is a natural product isolated or synthesized for the first time
We encourage authors reporting various compounds or compound libraries to apply the FAIR principles and include a summary file of these compounds as part of the submission. This file should be deposited in an appropriate repository or be provided as part of the Supplementary Information, and should adhere to the following:
- format - CSV (*.csv), TSV (*.tsv) or SDF (*.sdf)
- for chemical structures - relevant headers including SMILES , InChI and InChIKey
- for chemical names - Name and Synonym
- for other comments - such data, metadata, etc
These instructions are based on FAIR chemical structures in the Journal of Cheminformatics (E.L. Schymanski & E.E. Bolton, Journal of Cheminformatics , 2021, 13 , 50).
We recommend that authors follow the guidelines for the nomenclature of new radiolabelled compounds, as laid out in Consensus nomenclature rules for radiopharmaceutical chemistry - setting the record straight (C.H.H., G.A.D. et al., Nuclear Medicine and Biology , 2017, 55 , v – xi).
Authors should provide sufficient information to enable readers to reproduce any computational results. If software was used for calculations and is generally available, it should be properly cited in the references. References to the methods upon which the software is based should also be provided.
Equations, data, geometric parameters/coordinates, or other numerical parameters essential to the reproduction of the computational results (or adequate references when available in the open literature) should be provided. Authors who report the results of electronic structure calculations in relative energies should also include the absolute energies obtained directly from the computational output files. These may be deposited in an appropriate repository and cited, or provided in the Supplementary Information (SI).
We ask that the following information be provided where possible:
- As a minimum, papers reporting QM work should include the atomic coordinates, energies, and number of imaginary frequencies for all computed stationary points
- The level of theory used for computations should be mentioned in the text, and/or in the caption of the first figure that reports the results of those computations
- Where calculations are performed with density functional theory, the integration grid used for the calculations should be specified
- All relevant total energies (potential energies, enthalpies, Gibbs free energies, etc.) should be reported for each computed species. For transition states (TSs), the magnitude of the frequency of the imaginary vibrational mode may also be reported
- For intrinsic reaction coordinate (IRC) calculations, any shoulders in the IRC plots should be noted. If IRC calculations reveal that a TS corresponded to a reaction pathway different from that suggested by simple animation of its imaginary mode, then the IRC results (plots and details of reactants/products) should be deposited in an appropriate repository and cited
- Where computational work is included as part of a synthetic study, full details of any theoretical characterization (for example, computational NMR or VCD) of products and/or important synthetic intermediates should be documented
We also strongly encourage xyz, .mol2 or .pdb files for coordinates to be shared via deposition in an appropriate repository.
It is the responsibility of the authors to provide the raw data for all electrophoretic gel and blot data, ensuring sufficient evidence to support their conclusions.
All Western blot and other electrophoresis data should be supported by the underlying raw images. The image of the full gel and blot, uncropped and unprocessed, should be made available on submission. We suggest authors deposit their data in an appropriate repository. Where this isn’t possible, we ask authors to include the data as part of the Supplementary Information. All samples and controls used for a comparative analysis should be run on the same gel or blot.
When illustrating the result, any cropping or rearrangement of lanes within an image should be stated in the figure legend and with lane boundaries clearly delineated. Alterations should be kept to a minimum required for clarity.
Each image should be appropriately labelled, with the closest molecular mass markers and lanes labelled. All details should be visible; over or underexposed gels and blots are not acceptable. Authors should be able to provide raw data for all replicate experiments upon request.
Studies on fluorescence sensor systems should include titrations covering a full range of analyte concentration, from the absence of analyte to a stoichiometric excess, taking the following factors into account:
- If the analyte shows significant absorption at the excitation or emission frequencies corrections should be carried out for Inner Filter Effects (IFEs). (note: fluorescence probes where the response mechanism is based on the Inner Filter Effect are not suitable for publication in NJC)
- A plateau should be observed at high analyte concentrations for the intensity vs. concentration plot when the sensing mechanism is based on association
- Calculated Limits of Detection (LoD) should be supported by experimental data at similar concentrations
- The intensity vs. concentration relationship should be fitted using suitable software. The Benesi-Hildebrand linearization method for the determination of the association constant should not be used without extensive consideration of the limitations that arise from the method’s assumptions ( see Chemical Society Reviews Tutorial 10.1039/C0CS00062K for further details.)
Plots reporting the Stern-Volmer relationship (I°/I vs. concentration; the same should be valid for its reciprocal I/I°) should show an intercept of 1. Significant variation from this is not acceptable.
The Stern-Volmer relationship should be justified by reference to an appropriate quenching mechanism, e.g. dynamic quenching should show a linear relationship, while static quenching can present an upward curvature for relatively high association constants (see Chemical Society Reviews Tutorial 10.1039/D1CS00422K for further discussion)
The performance of all sensor systems should be compared to the current state-of-the-art sensors for the same analyte, with any differences in requirements (e.g. solvent system) clearly stated; a suitable (and justified) set of interferences should also be tested and discussed.
A new chemical substance (molecule or extended solid) should have a homogeneous composition and structure. Where the compound is molecular, authors should provide data to unequivocally establish its homogeneity, purity and identification as described above.
In general, this should include elemental analyses or a justification for the omission of this data.
This is particularly important for NMR silent paramagnetic compounds where NMR data tends to be less useful in establishing purity. In some instances an assigned 1 H NMR spectrum of a paramagnetic compound that is demonstrably devoid of impurities may be acceptable.
It may be possible to substitute elemental analyses with high-resolution mass spectrometric molecular weights. This is appropriate, for example, with trivial derivatives of thoroughly characterised substances or routine synthetic intermediates. In all cases, relevant spectroscopic data (NMR, IR, UV-vis, etc.) should be provided in tabulated form or as reproduced spectra. These may be deposited in an appropriate repository and cited, or provided in the Supplementary Information (SI). However, it should be noted that, in general, mass spectrometric and spectroscopic data do not constitute proof of purity, and, in the absence of elemental analyses, additional evidence of purity should be provided (melting points, PXRD data, etc.).
Where the compound is an extended solid, it is important to unequivocally establish the chemical structure and bulk composition. Single crystal X-ray diffraction does not determine the bulk structure. Reviewers will normally look to see evidence of bulk homogeneity. A fully indexed powder diffraction pattern that agrees with single crystal data may be used as evidence of a bulk homogeneous structure, and chemical analysis may be used to establish purity and homogeneous composition.
Detailed information on the reporting requirements for X-ray crystallography, including small molecule single crystal data and powder diffraction data is available in the section on X-ray crystallography.
Novel macromolecular structures and newly reported nucleic acid or protein sequences and microarray data should be deposited in appropriate repositories. It is the responsibility of the authors to provide relevant accession numbers prior to publication.
A Data Availability Statement with suitable links to the deposited data should be included. Please see our Data Sharing policy for more details. For high-throughput studies, we encourage authors to refer to Minimum Information Standards as determined and maintained by the relevant communities. For further details see:
- Minimum information standard - Wikipedia
- Minimum Information for Biological and Biomedical Investigations - FAIRsharing Information Resource
The following should be supplied for macromolecular X-ray structures:
- R merge , completeness, multiplicity and I /sigma(I) - both overall and in the outer resolution shell - for data, and
- R cryst , R free and the bond and angle deviations for coordinates
- a Ramachandran plot, and preferably
- real space R -factor
For NMR structures equivalent data plus resonance assignments should be supplied - number of restraints (NOEs and J -couplings), RMS restraint deviation, etc, plus resonance assignments should be supplied.
All the above information should be included as summary data tables in the manuscript or may be deposited in an appropriate repository and cited, or provided in the Supplementary Information.
Magnetic measurements
If data from magnetic measurements are presented, the authors should provide a thorough description of the experimental details pertaining to how the sample was measured. If the data have been corrected for sample or sample holder diamagnetism, the diamagnetic correction term should be provided and the manner in which it was determined should be stated.
Any fit of magnetic data (for example, χ(T), χ(1/T), χT(T), μ(T), M(H), etc.) to an analytical expression should be accompanied by the Hamiltonian from which the analytical expression is derived, the analytical expression itself, and the fitting parameters. If the expression is lengthy, it may be deposited in an appropriate repository and cited, or relegated to the Supplementary Information to conserve space. When an exchange coupling constant (J) is quoted in the abstract, the form of the Hamiltonian should also be included in the abstract.
For nanomaterials (such as quantum dots, nanoparticles, nanotubes, nanowires), it is the authors’ responsibility not only to provide a detailed characterisation of individual components (see Inorganic and organometallic compounds ) but also a comprehensive characterisation of the bulk composition. Characterisation of the bulk sample could require determination of the chemical composition and size distribution over large portions of the sample.
All nanoparticulate materials should have been purified from synthesis by-products and residual parent compounds, ions etc. If they are to be applied in dispersed form (for example, as a nanoparticulate drug carrier), sufficient data on the dispersion state should be provided (for example, by dynamic light scattering, centrifugal analysis, nanoparticle tracking analysis).
SEM or TEM images for hybrid inorganic-organic nanoparticles should be provided in at least three different levels of magnification. Bar scales should be clearly visible. Images may be deposited in an appropriate repository and cited, or provided in the Supplementary Information (SI).
It is the responsibility of the authors to provide unequivocal support for the purity and assigned structure of all compounds using a combination of the following characterisation techniques.
Elemental analysis is recommended to confirm sample purity and corroborate isomeric purity. We encourage authors to provide instrumental details and the chromatograms of the performed measurements in the Supplementary Information where possible. Authors are also requested to provide 1 H, 13 C NMR spectra and/or GC/HPLC traces if satisfactory elemental analysis cannot be obtained.
For libraries of compounds, HPLC traces should be submitted as proof of purity. The determination of enantiomeric excess of nonracemic, chiral substances should be supported with either GC/HPLC traces with retention times for both enantiomers and separation conditions (that is, chiral support, solvent and flow rate) or for Mosher Ester/Chiral Shift Reagent analysis, copies of the spectra.
Important physical properties, for example, boiling or melting point, specific rotation, refractive index, including conditions and a comparison to the literature for known compounds, should be provided. For crystalline compounds, the method used for recrystallisation should also be documented (that is, solvent etc.).
Mass spectra and a complete numerical listing of 1 H, 13 C NMR peaks in support of the assigned structure, including relevant 2D NMR and related experiments (that is, NOE, etc.) are required. As noted in Analytical , authors are requested to provide copies of these spectra. Infrared spectra that support functional group modifications, including other diagnostic assignments, should be included. High-resolution mass spectra are acceptable as proof of the molecular weight providing the purity of the sample has been accurately determined as outlined above.
For all soluble polymers, an estimation of molecular weight should be provided by size exclusion chromatography, including details of columns, eluents and calibration standards, intrinsic viscosity, MALDI TOF, etc. In addition, full NMR characterisation ( 1 H, 13 C) as for organic compound characterisation above.
For Gel Permeation Chromatography, molecular weight (Mw), molecular number (Mn) polydispersity index (PDI), and internal standards used should be specified, and associated images/spectra should be made available on submission. We suggest authors deposit their data in an appropriate repository. Where this isn’t possible, we ask authors to include the data as part of the article Supplementary Information (SI).
These should be described in enough detail so that a skilled researcher is able to repeat them. They should include the specific reagents, products and solvents with all of their amounts (g, mmol, for products: %), as well as clearly stating how the percentage yields are calculated.
- Reaction times, temperature and solvent quantities should be reported
- Reactions requiring heating - provide the heat source
- Reactions conducted using microwave heating - information on the type of vessel used and how the temperature was monitored should be given as well as the temperature reached or maintained
- Light-promoted reactions - report the light source and specific conditions
- Describe purification methods in detail
- GC or HPLC traces should be supported with retention times and separation conditions (support, solvent and flow rate)
- Centrifugation - includes rotation speed, centrifugation/dialysis time, solution for resuspension, resuspension time and procedure for each centrifugation step
Synthetic procedures should also include all the characterisation data for the prepared compound or material. For a series of related compounds at least one representative procedure that outlines a specific example that is described in the text or in a table and that is representative of the other cases should be provided. For a multistep synthesis, spectra of key compounds and the final product should be included.
Systems Biology Markup Language (SBML) is a computer-readable format for representing models of biochemical reaction networks. SBML is applicable to metabolic networks, cell-signalling pathways, regulatory networks, and many others.
We encourage authors to prepare models of biochemical reaction networks using SBML and to deposit the model in an open database such as the BioModels database or MetabolicAtlas .
Include some physical or experimental validation. Studies that screen a molecule against a set of receptors with no link to physical or experimental data are not suitable for publication.
These guidelines provide details for the presentation of single crystal and powder diffraction data; they apply to submissions to any of our journals.
Authors should present their crystal data in a CIF (Crystallographic Information File) format and deposit this with the Cambridge Crystallographic Data Centre (CCDC) before submission. Data will be held in the CCDC's confidential archive until publication of the article, but it will be made accessible to reviewers and the publisher assigned to review the data.
At the point of publication, any deposited data will be made publicly available through the CCDC Access Structures service. In addition, organic and metal-organic structures will be curated into the Cambridge Structural Database, and inorganic structures will be curated into the Inorganic Crystal Structures Database (FIZ Karlsruhe).
Upon deposition, each data set is assigned a Digital Object Identifier (DOI), so that the crystal structure is unambiguously identified and registered.
Include CCDC or ICSD numbers in the manuscript prior to submission as part of a Data Availability Statement . During submission authors will be asked to cite CCDC or ICSD reference numbers; CIFs should not be submitted with the manuscript. Any revised CIFs should be deposited directly with the CCDC before the revised manuscript is submitted to us.
In addition, authors are required to provide a checkCIF report for their reported crystal data. The checkCIF report can be obtained via the International Union of Crystallography's (IUCr) free checkCIF service , or as part of the CCDC deposition process. Any ‘level A' alerts in the report should be explained in the submission details for the article or an explanation provided within the submitted CIF. Authors should submit the checkCIF reports to the Royal Society of Chemistry along with the manuscript files.
If the editor deems it necessary during the peer-review process, the crystallography associated with the manuscript may undergo specialist crystallographic assessment, in which case a report will be provided along with the other reports from reviewers. Any points raised in this assessment should be attended to and all revised CIFs should be deposited with the CCDC prior to uploading the revised manuscript.
For recommended information to include in your CIF, please see the CCDC CIF Deposition Guidelines. If SQUEEZE or MASK procedures are used, this should be noted in the CIF file.
We encourage authors to include hkl data in the deposited CIF file. Alternatively authors can submit hkl data and the structure files (.fcf) separately during deposition with the CCDC. Raw data accompanying a structure should be made available by the authors for the review process, on request.
Details of the data collection and CCDC numbers should be given in the Data Availability Statement.
For relevant structures that are published as CSD communications, and that have not appeared in the manuscript or a previous journal publication, details should be included in the Data Availability Statement and the appropriate DOI should be cited as a reference in the manuscript.
Where there is significant discussion about the crystallography, the description may be given in textual or tabular form, although the latter is more appropriate if several structure determinations are being reported in one paper. A table of selected bond lengths and angles, with estimated standard deviations, should be restricted to significant dimensions only. Average values may be given with a range of E.S.D.s for chemically equivalent groups or for similar bonds.
Procedures for data collection and structure analysis can be provided as part of the Supplementary Information. The following data are recommended:
- Chemical formula and formula weight ( M )
- Crystal system
- Unit cell dimensions (Å or pm, degrees) and volume, with estimated standard deviations, temperature
- Space group symbol (if non-standard setting give related standard setting)
- Number of formula units in unit cell ( Z )
- Number of reflections measured and/or number of independent reflections, R int
- Final R values (and whether quoted for all or observed data)
- Flack or Rogers parameter (if appropriate)
For example:
Single crystals of [Pd{C(CO 2 Me)[C(CO 2 Me)C(CO 2 Me)=
C(CO 2 Me)C(CO 2 Me)=C(CO 2 Me)]C 6 H 3 [CH(Me)NH 2 ]-2-NO 2 -5}Br] 4 were recrystallised from dichloromethane, mounted in inert oil and transferred to the cold gas stream of the diffractometer.
Crystal structure determination of complex 4:
Crystal data. C 28 H 31 BrCl 4 N 2 O 14 Pd, M = 947.66, orthorhombic, a = 11.096(1), b = 17.197(2), c = 19.604(3) Å, U = 3741.0(9) Å 3 , T = 173 K, space group P2 1 2 1 2 1 (no.19), Z = 4, 6013 reflections measured, 5665 unique (R int = 0.031), which were used in all calculations. The final wR(F 2 ) was 0.099 (all data).
There may be cases where authors do not wish to include details or extensive discussion of a crystal structure determination. Examples include where only the connectivity has been established, data is marked as low quality at the CCDC, the structure is not integral to the conclusions of the article, or the structure has been discussed in a previous publication. Authors should be mindful of unnecessary fragmentation and the editor’s decision on this will be final.
Authors are encouraged to submit powder diffraction crystallographic data as a CIF (Crystallographic Information File) file to an appropriate repository. Please see the Data Sharing policy for further details.
Authors should combine multiple data sets for a given manuscript into a single file. The individual structures in the combined file should be separated from each other by the sequence #===END at the beginning of a line.
Authors should identify the manuscript with which the electronic file is associated when they submit the file by entering the name of the manuscript at the top of the electronic file.
The information required for deposition includes the following:
- A table of final fractional atomic coordinates
- Any calculated coordinates (for example, hydrogen)
- A full list of bond lengths and angles with estimated standard deviations
- A full list of displacement parameters in the form B ij or U ij (in Å 2 or pm 2 )
- Full details of the refinement
- Profile difference plots for all analyses. Where a range of similar analyses are presented a minimum number of representative plots may be given
Unrefined powder diffraction data should normally be reported only if the data form part of the discussion presented in the paper, and should be restricted to new materials. In such cases, the following experimental details should be provided in either textual or tabular format:
- Diffractometer name and model
- Radiation wavelength (Å)
- Temperature of data collection
- Unit cell dimensions (Å or pm, degrees), if determined
Tables of 2 θ data, or diagrams showing diffraction patterns of reaction products, should not normally be published in print unless they have some distinct feature of relevance that requires such detail to be present. In most cases, such data may be provided as Supplementary Information (SI).
For cases where the materials are new , but have similar powder data to other well-characterised materials, such data should not usually be included in the paper but can be deposited in an appropriate repository, with the relevant reference number included in a Data Availability Statement, and cited.
For refined powder diffraction data (where atomic coordinates have been determined), if the procedures for data collection and structure analysis were routine, their description may be concise. When the analysis has not been of a routine nature, the authors should briefly detail the procedures used. In most cases, a table of atomic coordinates may be provided, which should give details of occupancies that are less than unity.
Anisotropic thermal parameters may be included if they form an important aspect of the study. Selected bond lengths and angles, with estimated standard deviations, should be given.
For Rietveld refinements, an observed + calculated + difference profile plot should normally be given for each structure determination, except where a significant number of similar refinements have been carried out. In such cases, only the minimum number of representative plots should be included in the article, with additional plots being deposited in an appropriate repository and cited, or included in the SI.
The following information should be provided:
- Unit cell dimensions (Å or pm, degrees)
- Space group
- Number of formula units in unit cell ( Z )
- Number of reflections
- Final R values ( R wp , R exp and R l ) and method of background treatment
Data should still be deposited with the CCDC and the appropriate CCDC reference numbers and DOIs cited.
For information on how to cite crystallographic data in your manuscript, please see the section on Data Citation.
Submit your journal article
Data sharing guidance and policy, author guidelines and information.
← Explore all information and guidelines for authors

Presenting Data – Graphs and Tables
Types of data.
There are different types of data that can be collected in an experiment. Typically, we try to design experiments that collect objective, quantitative data.
Objective data is fact-based, measurable, and observable. This means that if two people made the same measurement with the same tool, they would get the same answer. The measurement is determined by the object that is being measured. The length of a worm measured with a ruler is an objective measurement. The observation that a chemical reaction in a test tube changed color is an objective measurement. Both of these are observable facts.
Subjective data is based on opinions, points of view, or emotional judgment. Subjective data might give two different answers when collected by two different people. The measurement is determined by the subject who is doing the measuring. Surveying people about which of two chemicals smells worse is a subjective measurement. Grading the quality of a presentation is a subjective measurement. Rating your relative happiness on a scale of 1-5 is a subjective measurement. All of these depend on the person who is making the observation – someone else might make these measurements differently.
Quantitative measurements gather numerical data. For example, measuring a worm as being 5cm in length is a quantitative measurement.
Qualitative measurements describe a quality, rather than a numerical value. Saying that one worm is longer than another worm is a qualitative measurement.
| Quantitative | Qualitative | |
| Objective | The chemical reaction has produced 5cm of bubbles. | The chemical reaction has produced a lot of bubbles. |
| Subjective | I give the amount of bubbles a score of 7 on a scale of 1-10. | I think the bubbles are pretty. |
After you have collected data in an experiment, you need to figure out the best way to present that data in a meaningful way. Depending on the type of data, and the story that you are trying to tell using that data, you may present your data in different ways.
Data Tables
The easiest way to organize data is by putting it into a data table. In most data tables, the independent variable (the variable that you are testing or changing on purpose) will be in the column to the left and the dependent variable(s) will be across the top of the table.
Be sure to:
- Label each row and column so that the table can be interpreted
- Include the units that are being used
- Add a descriptive caption for the table
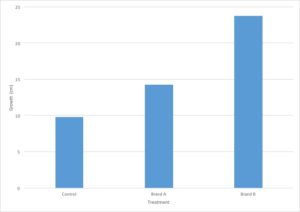
You are evaluating the effect of different types of fertilizers on plant growth. You plant 12 tomato plants and divide them into three groups, where each group contains four plants. To the first group, you do not add fertilizer and the plants are watered with plain water. The second and third groups are watered with two different brands of fertilizer. After three weeks, you measure the growth of each plant in centimeters and calculate the average growth for each type of fertilizer.
| Treatment | Plant Number | ||||
| 1 | 2 | 3 | 4 | Average | |
| No treatment | 10 | 12 | 8 | 9 | 9.75 |
| Brand A | 15 | 16 | 14 | 12 | 14.25 |
| Brand B | 22 | 25 | 21 | 27 | 23.75 |
Scientific Method Review: Can you identify the key parts of the scientific method from this experiment?
- Independent variable – Type of treatment (brand of fertilizer)
- Dependent variable – plant growth in cm
- Control group(s) – Plants treated with no fertilizer
- Experimental group(s) – Plants treated with different brands of fertilizer
Graphing data
Graphs are used to display data because it is easier to see trends in the data when it is displayed visually compared to when it is displayed numerically in a table. Complicated data can often be displayed and interpreted more easily in a graph format than in a data table.
In a graph, the X-axis runs horizontally (side to side) and the Y-axis runs vertically (up and down). Typically, the independent variable will be shown on the X axis and the dependent variable will be shown on the Y axis (just like you learned in math class!).
Line graphs are the best type of graph to use when you are displaying a change in something over a continuous range. For example, you could use a line graph to display a change in temperature over time. Time is a continuous variable because it can have any value between two given measurements. It is measured along a continuum. Between 1 minute and 2 minutes are an infinite number of values, such as 1.1 minute or 1.93456 minutes.
Changes in several different samples can be shown on the same graph by using lines that differ in color, symbol, etc.
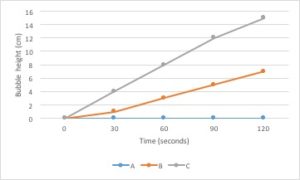
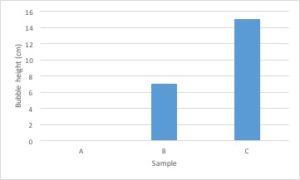
Bar graphs are used to compare measurements between different groups. Bar graphs should be used when your data is not continuous, but rather is divided into different categories. If you counted the number of birds of different species, each species of bird would be its own category. There is no value between “robin” and “eagle”, so this data is not continuous.
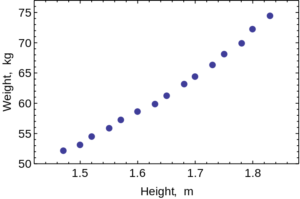
Scatter Plot
Scatter Plots are used to evaluate the relationship between two different continuous variables. These graphs compare changes in two different variables at once. For example, you could look at the relationship between height and weight. Both height and weight are continuous variables. You could not use a scatter plot to look at the relationship between number of children in a family and weight of each child because the number of children in a family is not a continuous variable: you can’t have 2.3 children in a family.
How to make a graph
- Identify your independent and dependent variables.
- Choose the correct type of graph by determining whether each variable is continuous or not.
- Determine the values that are going to go on the X and Y axis. If the values are continuous, they need to be evenly spaced based on the value.
- Label the X and Y axis, including units.
- Graph your data.
- Add a descriptive caption to your graph. Note that data tables are titled above the figure and graphs are captioned below the figure.
Let’s go back to the data from our fertilizer experiment and use it to make a graph. I’ve decided to graph only the average growth for the four plants because that is the most important piece of data. Including every single data point would make the graph very confusing.
- The independent variable is type of treatment and the dependent variable is plant growth (in cm).
- Type of treatment is not a continuous variable. There is no midpoint value between fertilizer brands (Brand A 1/2 doesn’t make sense). Plant growth is a continuous variable. It makes sense to sub-divide centimeters into smaller values. Since the independent variable is categorical and the dependent variable is continuous, this graph should be a bar graph.
- Plant growth (the dependent variable) should go on the Y axis and type of treatment (the independent variable) should go on the X axis.
- Notice that the values on the Y axis are continuous and evenly spaced. Each line represents an increase of 5cm.
- Notice that both the X and the Y axis have labels that include units (when required).
- Notice that the graph has a descriptive caption that allows the figure to stand alone without additional information given from the procedure: you know that this graph shows the average of the measurements taken from four tomato plants.
Descriptive captions
All figures that present data should stand alone – this means that you should be able to interpret the information contained in the figure without referring to anything else (such as the methods section of the paper). This means that all figures should have a descriptive caption that gives information about the independent and dependent variable. Another way to state this is that the caption should describe what you are testing and what you are measuring. A good starting point to developing a caption is “the effect of [the independent variable] on the [dependent variable].”
Here are some examples of good caption for figures:
- The effect of exercise on heart rate
- Growth rates of E. coli at different temperatures
- The relationship between heat shock time and transformation efficiency
Here are a few less effective captions:
- Heart rate and exercise
- Graph of E. coli temperature growth
- Table for experiment 1
Principles of Biology Copyright © 2017 by Lisa Bartee, Walter Shriner, and Catherine Creech is licensed under a Creative Commons Attribution 4.0 International License , except where otherwise noted.
Share This Book
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Korean J Anesthesiol
- v.74(2); 2021 Apr
The principles of presenting statistical results: Table
Sang gyu kwak.
1 Department of Medical Statistics, Daegu Catholic University School of Medicine, Daegu, Korea
2 Department of Anesthesiology and Pain Medicine, Chung-Ang University College of Medicine, Seoul, Korea
Jong Hae Kim
3 Department of Anesthesiology and Pain Medicine, Daegu Catholic University School of Medicine, Daegu, Korea
Tae Kyun Kim
4 Department of Anesthesiology and Pain Medicine, Yangsan Hospital, Pusan National University School of Medicine, Busan, Korea
Dong Kyu Lee
5 Department of Anesthesiology and Pain Medicine, Guro Hospital, Korea University School of Medicine, Seoul, Korea
Sangseok Lee
6 Department of Anesthesiology and Pain Medicine, Sanggye Paik Hospital, Inje University College of Medicine, Seoul, Korea
Jae Hong Park
7 Department of Anesthesiology and Pain Medicine, Haeundae Paik Hospital, Inje University College of Medicine, Busan, Korea
Francis Sahngun Nahm
8 Department of Anesthesiology and Pain Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
9 Department of Anesthesiology and Pain Medicine, Dongguk University Ilsan Hospital, Goyang, Korea
General medical journals such as the Korean Journal of Anesthesiology (KJA) receive numerous manuscripts every year. However, reviewers have noticed that the tables presented in various manuscripts have great diversity in their appearance, resulting in difficulties in the review and publication process. It might be due to the lack of clear written instructions regarding reporting of statistical results for authors. Therefore, the present article aims to briefly outline reporting methods for several table types, which are commonly used to present statistical results. We hope this article will serve as a guideline for reviewers as well as for authors, who wish to submit a manuscript to the KJA.
Introduction
It has been encouraging to see the growing number of outstanding article submissions and publications in the Korean Journal of Anesthesiology (KJA) over the years. Unfortunately, however, the diversity of result presentation format, alongside the number of manuscripts, has resulted in confusion not only in the review and publication process but also in delivering appropriate information to the readers. Presenting results derived using similar statistical methods in a prescribed tabular format recommended by the journal will not only simplify the review and publication process but also help with readers’ understanding of the published content.
General methods of presenting easy-to-read results can be found in the previous article [ 1 ]. In this article, we present specific examples of the appropriate application of the Instruction for Authors of KJA 1) to the tabular results for various analysis methods commonly used in research.
Common statistical tests and tables
Various types of tables are used to clearly present various forms of research results. Even if presented independently, tables must contain the essential elements needed to convey the necessary information. For instance, the title must contain sufficient description of the content, while the body and footnotes of the table must describe in detail the statistical method and results of the analysis.
The following are the examples of typical tabular results commonly submitted to this journal. The data used in the examples were generated randomly, unless otherwise indicated, and do not reflect results of a specific study, i.e., the presented results have no clinical significance.
Common guidelines
Statistic provided in all the tables follow the guidelines on the representation of significant figures and statistics in the Instructions for authors provided by the KJA. There must be no blank spaces in the table and estimate, sample size (n), and the statistical method used must be appropriately included when presenting results using statistical analysis. Quantitative data can be expressed as a representative value and its distribution, such as ‘mean ± standard deviation’ or ‘median (first quartile, third quartile). Qualitative data can be expressed as ‘frequency (percent, %)’, et al. For statistical analyses involving variable transformation that changes the shape of the distribution (i.e., log transformation), statistic reflective of the original value should be used. An inverse statistic may also be expressed if needed. Description of the transformation method and transformed values must accompany variable transformation. For more information on data transformation, refer to Lee's article [ 2 ].
Based on the statistic derived from the sample data of the study, the population parameters are estimated. It is recommended to display a confidence interval (i.e., 95% CI) that is an interval estimator along with point estimators such as mean, median, proportion, coefficient, et al. The previously derived estimates are described together with the hypothesis test results. P value must be described in three decimal places, and a test statistic should be presented in detail so that statistical inference can be made. Presenting the effect size, if possible, can aid the interpretation of statistical results.
An explanation of the abbreviations must be included in the footnote even if an explanation is provided in the text so that the table can be interpreted independently. The unit of measure of each variable must be accurately described, and the number of samples should be presented in the title or alongside the variable.
One sample comparison
In one sample comparisons, the data of the experimental sample are compared to a specific reference value. The example in Table 1 is a comparison between the arterial pressures of the experimental sample with the reference value of 60 mmHg. Based on the distribution of experimental data, a parametric or non-parametric method of statistical analysis was applied, along with the difference between the reference value and that of the experimental sample and its 95% CI.
Example of One Sample Comparison with Reference Value
| Variables | Results | Reference value | Difference (95% CI) | P value |
|---|---|---|---|---|
| MBP (mmHg, n = 30) | 70.0 ± 5.0 | 60 | 10.0 (8.0, 12.0) | < 0.001 |
| MBP (mmHg, n = 28) | 70 (64.0, 75.0) | 60 | 10.0 (8.0, 12.0) | < 0.001 |
Values are presented as mean ± SD or median (Q1, Q3). MBP: mean blood pressure.
* One-sample t-test,
† Two-sided P value < 0.05,
‡ Wilcoxon’s signed rank test. These values, including P values, are presented according to the Instructions for Authors of Korean Journal of Anesthesiology for notation below the decimal point.
In the case of categorical data, proportions, etc. can be compared. One sample proportion test can be performed to compare the response rate with the reference value, and when the response rate is close to 0% or 100%, an exact binomial test is sometimes performed. The comparison results are described in Table 2 along with the 95% CI of the response rate.
Example of One Sample Test of Proportions
| Variables | Positive response | Reference probability | Response rate (95% CI) | P value |
|---|---|---|---|---|
| PONV (n = 25) | 9 | 0.20 | 0.36 (0.18, 0.57) | 0.080 |
| Itching sense (n=64) | 5 | 0.02 | 0.08 (0.03, 0.17) | 0.009 |
PONV: postoperative nausea and vomiting,
* One sample proportion test with continuity correction,
† Exact binomial test,
‡ Two-sided P value < 0.05. These values, including P values, are presented according to the Instructions for Authors of Korean Journal of Anesthesiology for notation below the decimal point.
Comparison of two independent samples
Table 3 presents the results of comparing the mean arterial pressure and heart rate after endotracheal intubation when two types of endotracheal intubation devices were used. A parametric or non-parametric method of statistical analysis is applied depending on the distribution of the experimental data. The statistical method used is described in the table along with a representative value suitable for the distribution. To facilitate the interpretation of the results, the difference between the two groups is presented and the corresponding P value is presented to three decimal places.
Example of Independent Two Sample Comparison
| Variables | Group S (n = 49) | Group P (n = 53) | Difference (95% CI) | P value |
|---|---|---|---|---|
| MBP (mmHg) | 72.3 ± 14.3 | 73.1 ± 14.9 | −0.8 (−6.5, 4.9) | 0.781 |
| Heart rate | 89.0 (75.0, 103.0) | 82.0 (72.0, 93.0) | 7.0 (0, 14.0) | 0.062 |
* Independent two sample t-test,
† Mann-Whitney U test. These values, including P values, are presented according to the Instructions for Authors of Korean Journal of Anesthesiology for notation below the decimal point.
Comparison of matched pairs
Table 4 presents data from a study measuring mean blood pressure before and after the administration of a drug. The table presents results of administering the drug in a sample of hypertensive patients and a sample of patients with a body mass index over 30 kg/m 2 . The number of patients in each sample has been presented and the mean or median blood pressure was used as the representative value according to the distribution of measurements. A paired t-test was used to perform a paired comparison using the difference in pre- and post-treatment values for each patient. The statistically estimated differences are presented alongside the its 95% CI. The statistical method and P value is also clearly presented.
Example of Dependent Two Samples Comparison
| Underlying factors | MBP | Mean difference (95% CI) | P value | |
|---|---|---|---|---|
| Pre-treatment | Post-treatment | |||
| Hypertension (n = 20) | 74.0 ± 13.9 | 70.9 ± 13.6 | 3.1 (0.4, 5.8) | 0.026 |
| BMI > 30 kg/m (n = 25) | 75.4 (66.8, 81.5) | 73.9 (65.0, 84.5) | 1.5 (-1.0, 4.0) | 0.228 |
Values are presented as mean ± SD or median (Q1, Q3). BMI: body mass index, MBP: mean blood pressure.
* Paired t-test,
Comparison of three or more independent samples
Results from a study on pain control following a Cesarean section are presented in Table 5 [ 3 ]. The administered dose of morphine and time taken until the first dose was compared between a control group that received normal saline, a group that received intrathecal morphine and a group that received a quadratus lumborum block. Morphine requirement with a normal distribution was expressed as ‘mean ± standard deviation’, while the time taken until the first dose was expressed as a ‘median (Q1, Q3)’ value as it did not satisfy the normal distribution assumption. An accurate P value, up to 3 decimal places, and the number of samples in each group are presented, while a detailed description of the statistical method including the multiple comparison method for post hoc analysis is provided.
Example of Three Independent Samples Comparison
| Variables | Control group (n = 30) | ITM group (n = 30) | QLB group (n = 30) | P value |
|---|---|---|---|---|
| Morphine requirement (mg) | 61.0 ± 12.9 | 42.8 ± 10.4 | 18.2 ± 9.6 | <0.001 |
| Time to first morphine dose (h) | 2 (0.5, 4) | 8 (3, 24) | 17 (6, 36) | 0.002 |
Values are presented as mean ± SD or median (Q1, Q3). ITM: intrathecal morphine, QLB: quadratus lumborum block. P values indicate the statistical inference result of overall comparisons.
* One-way analysis of variance with Tukey’s method,
‡ Kruskal-Wallis H test with Dunn’s procedure. These values, including P values, are presented according to the Instructions for Authors of Korean Journal of Anesthesiology for notation below the decimal point. Excerpt from Salama ER [ 3 ] results showing representative values and P value as examples of comparison of three independent samples.
Categorical data comparison
Table 6 presents results of an investigation analyzing the occurrence of successful endotracheal intubation, sore throat 1 hour following tracheal extubation, and post-surgical vocal cord paralysis in two groups treated with an existing versus newly developed endotracheal tube. Results were reported as the frequency of occurrence and relative frequency and the statistical method and P value are clearly presented.
Example of Categorical Data Comparison
| Variables | Group N (n = 49) | Group C (n = 53) | P value |
|---|---|---|---|
| Successful tracheal intubation | 44 (89.8) | 32 (60.4) | 0.001 |
| Sore throat at 1 h | 11 (22.4) | 20 (37.8) | 0.144 |
| Vocal cord paralysis | 1 (2.0) | 2 (3.8) | >0.999 |
Values are presented as frequency (%).
* Chi-squared test,
‡ Fisher’s exact test. These values, including P values, are presented according to the Instructions for Authors of Korean Journal of Anesthesiology for notation below the decimal point.
Logistic regression analysis
The dependent variable is a nominal scale. This analysis method is widely used when selecting a meaningful variable among various explanatory variables and results are presented in terms of odds ratio, etc. Parts of results of a study published in the KJA is presented in Table 7 [ 4 ]. The study analyzed the risk factors of post-anesthesia emergence agitation in the recovery room. Variables with three or more components were converted into insignificant dummy variables to estimate the odds ratio. The table presents the odds ratio of referenced components and those converted into dummy variables alongside the 95% CI. A detailed description of the statistical methods used to select variables in the logistic regression analysis is also included.
Risk Factors of Emergence Agitation in the PACU (n = 158) [ 4 ]
| Variables | Odds ratio (95% CI) | P value |
|---|---|---|
| Marital status | ||
| Divorced | Reference | |
| Single | 0.16 (0.04, 0.64) | 0.009 |
| Married | 0.16 (0.04, 0.62) | 0.008 |
| Pre-existing ND | 6.78 (1.36, 33.80) | 0.020 |
| Gynecological surgery | 0.29 (0.12, 0.71) | 0.007 |
| Thoracic surgery | 0.23 (0.07, 0.80) | 0.021 |
| IO bleeding | 1.00 (1.00, 1.00) | 0.047 |
| IO morphine administration | 1.15 (1.03, 1.28) | 0.015 |
| Analgesic drugs in PACU | 2.99 (1.56, 5.73) | 0.001 |
The odds ratio of Marital status is estimated with non-weighted dummified variables. IO: intraoperative, ND: neurologic disorders, PACU: post-anesthesia care unit,
* Backward binary stepwise logistic regression,
† Two-sided P value < 0.05. These values, including P values, are presented according to the Instructions for Authors of Korean Journal of Anesthesiology for notation below the decimal point.
This article examined the principles of presenting the statistical results in clinical studies as a table. We hope to see manuscript submissions with standardized tables reflective of the provided framework. Such standardized format will help facilitate the submission and review process for both authors and reviewers.
1) https://ekja.org/authors/authors.php
Conflicts of Interest
All authors are Statistical Round Board Members in KJA.
Author Contributions
Sang Gyu Kwak (Conceptualization; Supervision; Writing – original draft; Writing – review & editing)
Hyun Kang (Validation; Writing – review & editing)
Jong Hae Kim (Validation; Writing – review & editing)
Tae Kyun Kim (Validation; Writing – review & editing)
EunJin Ahn (Validation; Writing – review & editing)
Dong Kyu Lee (Validation; Writing – original draft; Writing – review & editing)
Sangseok Lee (Project administration; Validation; Writing – review & editing)
Jae Hong Park (Validation; Writing – review & editing)
Francis Sahngun Nahm (Validation; Writing – review & editing)
Junyong In (Conceptualization; Supervision; Writing – original draft; Writing – review & editing)
| This web page presents a sample laboratory report written in a thermal fluids course (ME 2984) at Virginia Tech. Accompanying this report is a that states what the instructors expected as far as the scope of the experiment and the depth and organization of the report. |
| Voltage (V) | Voltage (V) | Pressure (kPa) | Temperature (K) | Temperature (K) |
|---|---|---|---|---|
| 6.32 | 0.0011 | 99.90 | 298.94 | 312.17 |
| 6.39 | 0.0020 | 102.81 | 320.32 | 321.28 |
| 6.78 | 0.0031 | 119.82 | 346.26 | 374.44 |
| 7.31 | 0.0046 | 145.04 | 381.64 | 453.24 |
| 7.17 | 0.0052 | 138.14 | 395.79 | 431.69 |
| 7.35 | 0.0064 | 147.04 | 424.09 | 459.50 |
| 7.45 | 0.0073 | 152.11 | 445.32 | 475.32 |
| 7.56 | 0.0078 | 157.78 | 457.11 | 493.04 |
| 7.66 | 0.0097 | 163.02 | 501.92 | 509.43 |
| 8.06 | 0.0107 | 184.86 | 525.51 | 577.69 |
| 8.10 | 0.0114 | 187.12 | 542.02 | 584.75 |
| 8.34 | 0.0130 | 200.97 | 579.75 | 628.03 |
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection

Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
Best practices for data management and sharing in experimental biomedical research
Affiliations.
- 1 Center for Neuroscience and Cell Biology, University of Coimbra, Coimbra, Portugal.
- 2 Center for Innovative Biomedicine and Biotechnology, University of Coimbra, Coimbra, Portugal.
- 3 Meta-Research Innovation Center at Stanford (METRICS), Stanford, California, United States.
- 4 Department of Statistics, Stanford University, Stanford, California, United States.
- PMID: 38451234
- DOI: 10.1152/physrev.00043.2023
Effective data management is crucial for scientific integrity and reproducibility, a cornerstone of scientific progress. Well-organized and well-documented data enable validation and building on results. Data management encompasses activities including organization, documentation, storage, sharing, and preservation. Robust data management establishes credibility, fostering trust within the scientific community and benefiting researchers' careers. In experimental biomedicine, comprehensive data management is vital due to the typically intricate protocols, extensive metadata, and large datasets. Low-throughput experiments, in particular, require careful management to address variations and errors in protocols and raw data quality. Transparent and accountable research practices rely on accurate documentation of procedures, data collection, and analysis methods. Proper data management ensures long-term preservation and accessibility of valuable datasets. Well-managed data can be revisited, contributing to cumulative knowledge and potential new discoveries. Publicly funded research has an added responsibility for transparency, resource allocation, and avoiding redundancy. Meeting funding agency expectations increasingly requires rigorous methodologies, adherence to standards, comprehensive documentation, and widespread sharing of data, code, and other auxiliary resources. This review provides critical insights into raw and processed data, metadata, high-throughput versus low-throughput datasets, a common language for documentation, experimental and reporting guidelines, efficient data management systems, sharing practices, and relevant repositories. We systematically present available resources and optimal practices for wide use by experimental biomedical researchers.
Keywords: biomedicine; data management; metadata; raw data; reporting guidelines; reproducibility.
PubMed Disclaimer
Similar articles
- A practical guide to data management and sharing for biomedical laboratory researchers. Fouad K, Vavrek R, Surles-Zeigler MC, Huie JR, Radabaugh HL, Gurkoff GG, Visser U, Grethe JS, Martone ME, Ferguson AR, Gensel JC, Torres-Espin A. Fouad K, et al. Exp Neurol. 2024 Aug;378:114815. doi: 10.1016/j.expneurol.2024.114815. Epub 2024 May 16. Exp Neurol. 2024. PMID: 38762093
- Management of Metadata Types in Basic Cardiological Research. Kusch H, Kossen R, Suhr M, Freckmann L, Weber L, Henke C, Lehmann C, Rheinländer S, Aschenbrandt G, Kühlborn LK, Marzec B, Menzel J, Schwappach B, Zelarayán LC, Cyganek L, Antonios G, Kohl T, Lehnart SE, Zoremba M, Sax U, Nussbeck SY. Kusch H, et al. Stud Health Technol Inform. 2021 Sep 21;283:59-68. doi: 10.3233/SHTI210542. Stud Health Technol Inform. 2021. PMID: 34545820
- A metadata schema for data objects in clinical research. Canham S, Ohmann C. Canham S, et al. Trials. 2016 Nov 24;17(1):557. doi: 10.1186/s13063-016-1686-5. Trials. 2016. PMID: 27881150 Free PMC article.
- Evaluation of repositories for sharing individual-participant data from clinical studies. Banzi R, Canham S, Kuchinke W, Krleza-Jeric K, Demotes-Mainard J, Ohmann C. Banzi R, et al. Trials. 2019 Mar 15;20(1):169. doi: 10.1186/s13063-019-3253-3. Trials. 2019. PMID: 30876434 Free PMC article. Review.
- Culture of Care: Organizational Responsibilities. Brown MJ, Symonowicz C, Medina LV, Bratcher NA, Buckmaster CA, Klein H, Anderson LC. Brown MJ, et al. In: Weichbrod RH, Thompson GA, Norton JN, editors. Management of Animal Care and Use Programs in Research, Education, and Testing. 2nd edition. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 2. In: Weichbrod RH, Thompson GA, Norton JN, editors. Management of Animal Care and Use Programs in Research, Education, and Testing. 2nd edition. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 2. PMID: 29787190 Free Books & Documents. Review.
Publication types
- Search in MeSH
Related information
Grants and funding.
- 101087416/EU Horizon
- PTDC/BTM-SAL/29297/2017,POCI-01-0145-FEDER-029297,UIDB/04539/2020,UIDP/04539/2020 and LA/P/0058/2/FTC Portugal
LinkOut - more resources
Full text sources.
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Experimental and theoretical study on anchorage loss of prestressed cfrp-reinforced concrete beams, 1. introduction, 2. fem calculation and theoretical analysis, 2.1. mechanism of anchorage loss, 2.2. description fem model, 2.3. analysis of fem results, 2.4. theoretical calculation analysis, 3. experimental study, 3.1. overview of project, 3.2. results and discussion, 3.3. theoretical validation, 4. conclusions.
- The FEM simulations revealed that the deformation of the connecting screw and the front end of the steel reaction frame accounts for approximately 95% of the total anchorage loss in a CFRP tendon–main beam connection system. The TCS was established based on the stress forms of these components, and the calculated results are consistent with the FEM analysis, indicating a high level of reliability.
- In the control group experiment, a CFRP tendon with a length of 12 m and a diameter of 12 mm, subjected to a prestress of 950 MPa, showed an anchorage loss of approximately 1.5% of the set prestress value, corresponding to an anchorage deformation of about 1 mm. Further studies indicated that the deformation magnitude is influenced by the prestress level, the dimensions of the steel reaction frame’s front end, the connecting screw length, and the thread gap values.
- The experimental results show relative discrepancies with both the FEM and theoretical results, attributed to gaps values ω 3 between the threaded connections. A 2 mm gap at each threaded connection was identified, and this can be reduced by tightening the threads with external force. However, the influencing pattern of these gaps requires further investigation. After adjusting the calculation formulas, the error was controlled within approximately 5%, demonstrating higher precision.
- This study provides a comprehensive framework for accurately predicting and mitigating anchorage loss in externally prestressed CFRP tendons, with significant implications for future engineering applications.
Author Contributions
Institutional review board statement, informed consent statement, data availability statement, conflicts of interest.
- Xu, L.; Xu, F. Design and Experimental Study on the External Reinforcement of Railway Prestressed Concrete Simply-supported Beam Bridges with CFRP Bars. Eng. Mech. 2013 , 30 , 89–95. [ Google Scholar ] [ CrossRef ]
- Najaf, E.; Orouji, M.; Ghouchani, K. Finite Element Analysis of the Effect of Type, Number, and Installation Angle of FRP Sheets on Improving the Flexural Strength of Concrete Beams. Case Stud. Constr. Mater. 2022 , 17 , e01670. [ Google Scholar ] [ CrossRef ]
- Hiroshi, F. FRP Composites in Japan. Concr. Int. 1999 , 21 , 29–32. [ Google Scholar ]
- Elrefai, A.; West, J.; Soudki, K. Fatigue of Reinforced Concrete Beams Strengthened with Externally Post-tensioned CFRP Tendons. Constr. Build. Mater. 2012 , 29 , 246–256. [ Google Scholar ] [ CrossRef ]
- Heydarinouri, H.; Nussbaumer, A.; Motavalli, M. Strengthening of Steel Connections in a 92-Year-Old Railway Bridge Using Pre-stressed CFRP Rods: Multiaxial Fatigue Design Criterion. J. Bridge Eng. 2021 , 26 , 04021023. [ Google Scholar ] [ CrossRef ]
- National Standard of the People’s Republic of China GB50010-2010 ; [2015 Edition] Code for Design of Concrete Structures. In Research Committee on Concrete Structures. China Planning Press: Beijing, China, 2015.
- Abass, B.; Mark, F.G.; Campbell, T.I. Fatigue Behavior of Concrete Beams Post-tensioned with Unbonded Carbon Fiber Reinforced Polymer Tendons. Can. J. Civ. Eng. 2006 , 33 , 1140–1155. [ Google Scholar ] [ CrossRef ]
- Kim, S.-H.; Park, S.Y.; Kim, S.T.; Jeon, S.-J. Analysis of Short-Term Prestress Losses in Post-tensioned Structures Using Smart Strands. Int. J. Concr. Struct. Mater. 2022 , 16 , 1. [ Google Scholar ] [ CrossRef ]
- Zhuge, P.; Zhiyu, J.; Zhanga, Z.; Ding, Y.; Hou, S. The influence of load transfer medium creep on the load-carrying capacity of the bond-type anchors of CFRP tendons. Constr. Build. Mater. 2019 , 206 , 236–247. [ Google Scholar ] [ CrossRef ]
- Cai, D.S.; Xu, Z.H.; Yin, J.; Liu, R.G.; Liang, G. A Numerical Investigation on the Performance of Composite Anchors for CFRP Tendons. Constr. Build. Mater. 2016 , 112 , 848–855. [ Google Scholar ] [ CrossRef ]
- Zhuge, P.; Yu, Y.; Cai, C.S.; Zhang, Z.; Ding, Y. Mechanical behavior and optimal design method for innovative CFRP cable anchor. J. Compos. Constr. 2019 , 23 , 04018067. [ Google Scholar ] [ CrossRef ]
- Mei, K.H.; Seracino, R.; Lv, Z.T. An Experimental Study on Bond Type Anchorages for Carbon Fiber-reinforced Polymer Cables. Constr. Build. Mater. 2016 , 106 , 584–591. [ Google Scholar ] [ CrossRef ]
- Xie, G.; Liu, R.; Chen, B. Synergistic Effect of a new wedge-bond-type anchor for CFRP tendons. J. Cent. South Univ. 2015 , 22 , 2260–2266. [ Google Scholar ] [ CrossRef ]
- Schmidt, J.W.; Bennitz, A.; Täljsten, B.; Pedersen, H. Development of mechanical anchor for CFRP tendons using integrated sleeve. J. Compos. Constr. 2010 , 14 , 397–405. [ Google Scholar ] [ CrossRef ]
- Zhou, J.; Wang, X.; Liu, X.; Shi, J.; Huang, H.; Li, Y.; Wu, Z. Numerical and experimental evaluation of a variable-stiffness wedge anchorage for basalt-fiber-reinforced polymer tendons. Eng. Struct. 2024 , 304 , 117684. [ Google Scholar ] [ CrossRef ]
- Páez, P.M. A simplified approach to determine the prestress loss and time-dependent deflection in cracked prestressed concrete members, prestressed with fiber reinforced polymers or steel tendons. Eng. Struct. 2023 , 279 , 115523. [ Google Scholar ] [ CrossRef ]
- Kim, Y.J.; Kang, J.-Y.; Park, J.-S.; Jung, W.-T. Prestress Loss of Post-Tensioned Near-Surface-Mounted Carbon Fiber-Reinforced Polymer for Bridge Strengthening. ACI Struct. J. 2018 , 115 , 1495–1506. [ Google Scholar ] [ CrossRef ]
- Wang, Z.; Du, X.; Liu, J. Studies on prestressing loss of concrete beams prestressed with CFRP tendons. Ind. Constr. 2011 , 41 , 29–32. [ Google Scholar ] [ CrossRef ]
- Xu, F. Study on Long-term Prestress Loss of Concrete Beams Strengthened by External Prestressing of CFRP Bar. Ind. Constr. 2017 , 47 , 248–250. [ Google Scholar ]
- Tao, G.; Zhuge, P.; Lin, D.; Zeng, T. Study on short-term prestress loss of bridge reinforced with large diameter carbon fiber bars. IOP Conf. Ser. Earth Environ. Sci. 2021 , 820 , 012001. [ Google Scholar ] [ CrossRef ]
- JSCE-CES 23. Recommendation for Design and Construction of Concrete Structures Using Continuous Fiber Reinforcing Materials , 1st ed.; Research Committee on Continuous Fiber Reinforcing Materials: Tokyo, Japan, 1997. [ Google Scholar ]
- Xu, X.; Ai, J.; Pan, J.; Cai, C. Experimental research on strengthened beam of reinforced concrete with prestressed FRP sheets and prestress loss. Ind. Constr. 2013 , 43 , 129–132. [ Google Scholar ]
- Deng, L.; Zhao, S.; Liao, L.; Yu, Z.; Ji, S. Study of prestress loss of steel structure strengthened with prestressed CFRP. Ind. Constr. 2014 , 44 , 147–150. [ Google Scholar ] [ CrossRef ]
- Wang, W.-W.; Dai, J.-G.; Harries, K.A.; Bao, Q.-H. Prestress Losses and Flexural Behavior of Reinforced Concrete Beams Strengthened with Posttensioned CFRP Sheets. J. Compos. Constr. 2012 , 16 , 207–216. [ Google Scholar ] [ CrossRef ]
- Kim, J.Y. Bond and Short-term Prestress Losses of Prestressed Composites for Strengthening PC Beams with Integrated Anchorage. J. Reinf. Plast. Compos. 2010 , 29 , 1277–1294. [ Google Scholar ] [ CrossRef ]
- Deng, J.; Rashid, K.; Li, X.; Xie, Y.; Chen, S. Comparative study on prestress loss and flexural performance of rectangular and T beam strengthened by prestressing CFRP plate. Compos. Struct. 2020 , 262 , 113340. [ Google Scholar ] [ CrossRef ]
- Shi, J.; Wang, X.; Wu, Z.; Zhu, Z. Optimization of anchorage and deviator for concrete beams prestressed with external fiber-reinforced polymer tendons. Compos. Struct. 2022 , 297 , 115970. [ Google Scholar ] [ CrossRef ]
Click here to enlarge figure
| Component | Material | Elastic Modulus (MPa) | Poisson Ratio |
|---|---|---|---|
| Steel reaction frame | Q235 | 210,000 | 0.2 |
| Fixed nut | Grade 8.8 | 210,000 | 0.3 |
| Connecting screw | Grade 8.8 | 210,000 | 0.3 |
| Anchor | Q235 | 210,000 | 0.3 |
| CFRP tendon | CFRP | 160,000 | 0.31 |
| Component | Deformation (mm) | Proportion of Total Deformation (%) | |
|---|---|---|---|
| Front end of the steel reaction frame | 0.270822 | 41.09 | |
| Fixed nut | 0.001575 | 0.24 | |
| Connecting screws | (a) | 0.027201 | 54.87 |
| (b) | 0.309988 | ||
| (c) | 0.024496 | ||
| Rear of anchor | 0.025051 | 3.8 | |
| Total deformation | 0.659133 | / | |
| Group | CFRP Tendon Length (mm) | Prestress (MPa) | Connecting Screw Length (mm) | Side Panel Spacing (mm) | Fixed Nut Torque (N·m) |
|---|---|---|---|---|---|
| 1 | 12,000 | 950 | 139 | 79 | / |
| 2 | 12,000 | 950 | 135 | 59 | 400 |
| 3 | 12,000 | 950 | 137 | 59 | / |
| 4 | 12,000 | 475 | 374 | 59 | 400 |
| 5 | 12,000 | 475 | 370 | 59 | / |
| 6 | 12,000 | 950 | 374 | 59 | 400 |
| 7 | 12,000 | 950 | 376 | 59 | / |
| Group | (MPa) | (MPa) | (MPa) | Deformation (a/mm) |
|---|---|---|---|---|
| 1 | 948.51 | 933.88 | 14.63 | 1.061 |
| 2 | 951.52 | 941.65 | 9.87 | 0.716 |
| 3 | 947.98 | 935.71 | 12.27 | 0.890 |
| 4 | 486.86 | 476.28 | 10.58 | 0.784 |
| 5 | 485.58 | 472.43 | 13.16 | 0.954 |
| 6 | 975.28 | 957.20 | 18.08 | 1.311 |
| 7 | 962.36 | 941.14 | 21.22 | 1.539 |
| Group | Steel Reaction Frame | Connection Screw | Deformation | (MPa) |
|---|---|---|---|---|
| 1 | 0.267 | 0.364 | 0.631 | 8.70 |
| 2 | 0.112 | 0.355 | 0.467 | 9.196 |
| 3 | 0.111 | 0.359 | 0.470 | 11.995 |
| 4 | 0.057 | 0.471 | 0.528 | 10.046 |
| 5 | 0.057 | 0.465 | 0.522 | 12.719 |
| 6 | 0.118 | 0.978 | 1.097 | 17.888 |
| 7 | 0.113 | 0.936 | 1.049 | 19.988 |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Liu, Q.; Jiang, H.; Tao, G.; Zhuge, P. Experimental and Theoretical Study on Anchorage Loss of Prestressed CFRP-Reinforced Concrete Beams. Appl. Sci. 2024 , 14 , 6246. https://doi.org/10.3390/app14146246
Liu Q, Jiang H, Tao G, Zhuge P. Experimental and Theoretical Study on Anchorage Loss of Prestressed CFRP-Reinforced Concrete Beams. Applied Sciences . 2024; 14(14):6246. https://doi.org/10.3390/app14146246
Liu, Qinrui, Haozhe Jiang, Guocheng Tao, and Ping Zhuge. 2024. "Experimental and Theoretical Study on Anchorage Loss of Prestressed CFRP-Reinforced Concrete Beams" Applied Sciences 14, no. 14: 6246. https://doi.org/10.3390/app14146246
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals
Report Writing
Cite this chapter.

- R. H. Leaver 2 &
- T. R. Thomas 2
12 Accesses
Reports may vary considerably in form depending upon the circumstances of the writer. The researcher working in a university or college will publish his findings in order to inform and, hopefully, to gain the approbation of colleagues with similar interests. In an industrial setting his work is likely to have a more restricted circulation. In all cases the prime aim of the writer should be the concise and unambiguous communication of information.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Institutional subscriptions
Unable to display preview. Download preview PDF.
Erstwhile, (1968), Cylinder machining , Springer-Verlag, Berlin.
Google Scholar
Glaswegian, G. G. (1971), Proc. R.Soc. A293 .
Brown et al . (1972), Theory and design of copper cylinder. Trans. ASME 89F , 477–581.
Brown, T. A., Smith, J. A. and Jones, L. (1972) Theory and design of copper cylinder. Trans. Am. Soc. mech. Engrs 89F , 477–581.
Erstwhile, K. M. (1968) Maschinen-fabriken das Cylinders Springer-Verlag, Berlin.
Glaswegian, G. G. (1971) Cylinder turning: towards a new theory of centrifugal-force. Proc. R. Soc. A293 , 38–147.
D. H. Menzel, H. M. Jones and L. G. Boyd, Writing a Technical Paper , McGraw-Hill, New York, 1961.
B. M. Cooper, Writing Technical Reports , Penguin, London, 1964.
E. Gowers, The Complete Plain Words , Penguin, London, 1962.
H. W. Fowler, A Dictionary of Modern English Usage , 2nd edition, Oxford University Press, London, 1965.
Download references
Author information
Authors and affiliations.
Teesside Polytechnic, UK
R. H. Leaver ( Principal Lecturer in Mechanical Engineering ) & T. R. Thomas ( Senior Lecturer in Mechanical Engineering )
You can also search for this author in PubMed Google Scholar
Copyright information
© 1974 R. H. Leaver and T. R. Thomas
About this chapter
Leaver, R.H., Thomas, T.R. (1974). Report Writing. In: Analysis and Presentation of Experimental Results. Palgrave, London. https://doi.org/10.1007/978-1-349-01942-7_2
Download citation
DOI : https://doi.org/10.1007/978-1-349-01942-7_2
Publisher Name : Palgrave, London
Print ISBN : 978-0-333-14987-4
Online ISBN : 978-1-349-01942-7
eBook Packages : Palgrave Social & Cultural Studies Collection Social Sciences (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research

- Advanced Search
EduCross: : Dual adversarial bipartite hypergraph learning for cross-modal retrieval in multimodal educational slides
New citation alert added.
This alert has been successfully added and will be sent to:
You will be notified whenever a record that you have chosen has been cited.
To manage your alert preferences, click on the button below.
New Citation Alert!
Please log in to your account
Information & Contributors
Bibliometrics & citations, view options, recommendations, boosting deep cross-modal retrieval hashing with adversarially robust training.
Deep hashing methods effectively enhance the performance of conventional machine learning retrieval models, particularly in visual medium evolving cross-modal retrieval tasks, by relying on the outstanding feature extraction ability of deep neural ...
CM-GANs: Cross-modal Generative Adversarial Networks for Common Representation Learning
It is known that the inconsistent distributions and representations of different modalities, such as image and text, cause the heterogeneity gap, which makes it very challenging to correlate heterogeneous data and measure their similarities. Recently, ...
Deep adversarial metric learning for cross-modal retrieval
Cross-modal retrieval has become a highlighted research topic, to provide flexible retrieval experience across multimedia data such as image, video, text and audio. The core of existing cross-modal retrieval approaches is to narrow down the gap between ...
Information
Published in.
Elsevier Science Publishers B. V.
Netherlands
Publication History
Author tags.
- Dual generative adversarial network
- Cross-modal retrieval
- Deep bipartite hypergraph learning
- Framelet transform
- Multimodal educational slides
- Research-article
Contributors
Other metrics, bibliometrics, article metrics.
- 0 Total Citations
- 0 Total Downloads
- Downloads (Last 12 months) 0
- Downloads (Last 6 weeks) 0
View options
Login options.
Check if you have access through your login credentials or your institution to get full access on this article.
Full Access
Share this publication link.
Copying failed.
Share on social media
Affiliations, export citations.
- Please download or close your previous search result export first before starting a new bulk export. Preview is not available. By clicking download, a status dialog will open to start the export process. The process may take a few minutes but once it finishes a file will be downloadable from your browser. You may continue to browse the DL while the export process is in progress. Download
- Download citation
- Copy citation
We are preparing your search results for download ...
We will inform you here when the file is ready.
Your file of search results citations is now ready.
Your search export query has expired. Please try again.
Last updated 10th July 2024: Online ordering is currently unavailable due to technical issues. We apologise for any delays responding to customers while we resolve this. For further updates please visit our website https://www.cambridge.org/news-and-insights/technical-incident
We use cookies to distinguish you from other users and to provide you with a better experience on our websites. Close this message to accept cookies or find out how to manage your cookie settings .
Login Alert

- > A Student's Guide to Data and Error Analysis
- > Processing of experimental data

Book contents
- Frontmatter
- Part I Data and error analysis
- 1 Introduction
- 2 The presentation of physical quantities with their inaccuracies
- 3 Errors: classification and propagation
- 4 Probability distributions
- 5 Processing of experimental data
- 6 Graphical handling of data with errors
- 7 Fitting functions to data
- 8 Back to Bayes: knowledge as a probability distribution
- Answers to exercises
- Part II Appendices
- Part III Python codes
- Part IV Scientific data
5 - Processing of experimental data
Published online by Cambridge University Press: 05 June 2012
Access options
Save book to kindle.
To save this book to your Kindle, first ensure [email protected] is added to your Approved Personal Document E-mail List under your Personal Document Settings on the Manage Your Content and Devices page of your Amazon account. Then enter the ‘name’ part of your Kindle email address below. Find out more about saving to your Kindle .
Note you can select to save to either the @free.kindle.com or @kindle.com variations. ‘@free.kindle.com’ emails are free but can only be saved to your device when it is connected to wi-fi. ‘@kindle.com’ emails can be delivered even when you are not connected to wi-fi, but note that service fees apply.
Find out more about the Kindle Personal Document Service .
- Processing of experimental data
- Herman J. C. Berendsen , Rijksuniversiteit Groningen, The Netherlands
- Book: A Student's Guide to Data and Error Analysis
- Online publication: 05 June 2012
- Chapter DOI: https://doi.org/10.1017/CBO9780511921247.006
Save book to Dropbox
To save content items to your account, please confirm that you agree to abide by our usage policies. If this is the first time you use this feature, you will be asked to authorise Cambridge Core to connect with your account. Find out more about saving content to Dropbox .
Save book to Google Drive
To save content items to your account, please confirm that you agree to abide by our usage policies. If this is the first time you use this feature, you will be asked to authorise Cambridge Core to connect with your account. Find out more about saving content to Google Drive .
- Biochemistry and Molecular Biology
- Biostatistics
- Environmental Health and Engineering
- Epidemiology
- Health Policy and Management
- Health, Behavior and Society
- International Health
- Mental Health
- Molecular Microbiology and Immunology
- Population, Family and Reproductive Health
- Program Finder
- Admissions Services
- Course Directory
- Academic Calendar
- Hybrid Campus
- Lecture Series
- Convocation
- Strategy and Development
- Implementation and Impact
- Integrity and Oversight
- In the School
- In the Field
- In Baltimore
- Resources for Practitioners
- Articles & News Releases
- In The News
- Statements & Announcements
- At a Glance
- Student Life
- Strategic Priorities
- Inclusion, Diversity, Anti-Racism, and Equity (IDARE)
- What is Public Health?
Alumni Spotlight: Ruthe Huang, ScM '19
Ruthe Huang, ScM '19, is a Senior Data Scientist with Practical Intelligence, specializing in statistical modeling and data visualization.
Ruthe Huang, ScM '19, works for Practical Intelligence as a Senior Data Scientist, where she specializes in statistical modeling and data visualization. Her thesis, under the direction of Mei-Cheng Wang , was entitled "ADHD Risk Prediction in the Boston Birth Cohort: an Application of the Proportional Odds Model and ROC Curve Analysis.”
In her spare time, Huang volunteers for the Prison-to-Professionals program , tutoring math, statistics, and standardized test prep to criminal-justice involved groups. 2024 marks her seventh year working with the organization as their SAT/GRE Facilitator.
Describe your current position and responsibilities in a way that will inform current and prospective students about career opportunities in biostatistics.
I am a data scientist within the government contracting world, meaning that each client and problem set is never the same. For the last four years, I have been working for a Department of Defense client with uniquely frustrating data problems, including naïve and nontraditional sources of data, mission limitations to software and data access, and stakeholder distrust in statistical results. Much of my daily work involves not just data cleaning and statistical analysis but also working with software teams to deploy data products as well as presenting analytical results for a variety of audiences.
What sparked your interest in the field of biostatistics?
Early on in my undergraduate studies, I looked for a career field that would combine my two interests of public health and statistics; biostatistics was the perfect match. I loved that biostatistics was interdisciplinary, allowing me to master quantitative methods while still working in the realm of public health.
What led you to join the Johns Hopkins Department of Biostatistics?
I was biased in my decision, since I was a Hopkins undergrad and had the chance to experience how wonderful the faculty were before starting at Bloomberg. However, the ultimate decision came down to the small cohorts and robust statistical training that the Department offered.
I still find it challenging to convince stakeholders who are not familiar with data or quantitative methods to believe in the results of statistical modeling, but Dr. Zeger’s courses gave me the tools to continue improving my skills.
How did your degree prepare you for your career? What aspects of the Hopkins Biostatistics program did you find most useful?
The coursework of the ScM program took us through such rigorous detail that I left with the skillsets – and, more importantly, the confidence – to not fear strange or new data problems. In my career, I often face unorthodox problems with messy data; although I may not have learned how to deal with these problem sets through coursework, I have been able to apply foundational knowledge from our program to help clients conduct as robust of an analysis as possible or even help clients develop novel approaches.
In his Methods series, Dr. Zeger would often emphasize the importance of interpreting model results clearly and accurately. I think of this often in my career, as I find most of my presentations directed to (statistically) non-technical audiences. I still find it challenging to convince stakeholders who are not familiar with data or quantitative methods to believe in the results of statistical modeling, but Dr. Zeger’s courses gave me the tools to continue improving my skills.
What has been your most satisfying job experience using your biostatistics background?
It is often said that building data models is an art, not a science. I experienced this first-hand when I was able to identify a key result in data using only contextual knowledge and basic model-building techniques. A different organization analyzed the same dataset and used only machine learning algorithms. The other organization was not able to identify the key result from my simple classification model because they had not taken the time to understand the context from which the data came nor check the data assumption of using certain models.
How did you build your sense of community during your time as a Biostatistics student?
My cohort was the best part of my time as a Biostatistics student. Because we were a small group of six, we were able to spend a lot of time together both at Bloomberg as well as in our personal time. From working on assignments in our office to picking apples in Baltimore County, we were able to be a community of support for each other.
What was your favorite thing about living in Baltimore when you were a student?
My favorite thing about living in Baltimore as a student was exploring all the little pleasures the city has to offer. I always loved that Baltimore was a relatively small city, because it meant that there were less crowds to navigate through while enjoy the beautiful magnolia blossoms by the Washington Monument , eating the whole menu at Ekiben , or sweating your body weight at Artscape . Baltimore is beautiful in its juxtaposition: of loud and quiet, of dark and light, of chaotic and beautiful. MORE ALUMNI SPOTLIGHTS
Home Bancorp, Inc. 2024 Q2 - Results - Earnings Call Presentation
The following slide deck was published by Home Bancorp, Inc. in conjunction with their 2024 Q2 earnings call.

This article was written by
Recommended For You
About hbcp stock.
| Symbol | Last Price | % Chg |
|---|
More on HBCP
Trending analysis, trending news.

IMAGES
COMMENTS
Statements. The most common way of presentation of data is in the form of statements. This works best for simple observations, such as: "When viewed by light microscopy, all of the cells appeared dead." When data are more quantitative, such as- "7 out of 10 cells were dead", a table is the preferred form. Tables.
A "methods" section should include all the information necessary for someone else to recreate your experiment. Your experimental notes will be very useful for this section of the report. More or less, this section will resemble a recipe for your experiment. Don't concern yourself with writing clever, engaging prose.
Tables and figures are commonly adopted methods for presenting specific data or statistical analysis results. Figures can be used to display characteristics and distributions of data, allowing for intuitive understanding through visualization and thus making it easier to interpret the statistical results. To maximize the positive aspects of ...
This book is intended as a guide to the analysis and presentation of experimental results. It develops various techniques for the numerical processing of experimental data, using basic statistical methods and the theory of errors. After presenting basic theoretical concepts, the book describes the methods by which the results can be presented ...
Preface. This book is written as a guide for the presentation of experimental including a consistent treatment of experimental errors and inaccuracies. is meant for experimentalists in physics, astronomy, chemistry, life and engineering. However, it can be equally useful for theoreticians produce simulation data: they are often confronted with ...
Abstract. It important to properly collect, code, clean and edit the data before interpreting and displaying the research results. Computers play a major role in different phases of research starting from conceptual, design and planning, data collection, data analysis and research publication phases. The main objective of data display is to ...
Experimental Methods for Science and Engineering StudentsResponding to the developments of the past 20 years, Les Kirkup has thoroughly revised his popular book on experimental methods, while retaining the exten. ive coverage and practical advice from the first edition. Many topics from that edition remain, including documenting experiments ...
This book is intended as a guide to the analysis and presentation of experimental results. It develops various techniques for the numerical processing of experimental data, using basic statistical methods and the theory of errors. After presenting basic theoretical concepts, the book describes the methods by which the results can be presented, both numerically and graphically.
The chapter also considers how to express data, for example using scientific notation. The importance of presenting data clearly in tables is emphasised. In some situations, it is possible to estimate the size of values likely to emerge from an experiment, prompting our attention to be alerted when the values obtained differ considerably from ...
A central part of reporting any scientific work is the presentation of the results, in either tabular or, more commonly, graphical form, and a considerable literature has accumulated about the appropriate ways for displaying quantitative data (e.g. Cleveland 1993, Tufte 1983, 1990, and some recent issues of The American Statistician).Much of this literature focuses on clarity of graphs and it ...
This book is intended as a guide to the analysis and presentation of experimental results. It develops various techniques for the numerical processing of experimental data, using basic statistical methods and the theory of errors. After presenting basic theoretical concepts, the book describes the methods by which the results can be presented ...
Analysis and Presentation of Experimental Results. pp.301-375. The method of least squares is developed. The application of the method in the fitting of the best straight line or curve to a series ...
The following information is a guide to the presentation of experimental data, including appropriate formats for citation. Yield. Yield should be presented in parentheses after the compound name (or its equivalent). Weight and percentage should be separated by a comma - for example, the lactone (7.1 g, 56%). Melting point
Graphs are a powerful and concise way to communicate information. Representing data from an experiment in the form of an x-y graph allows relationships to be examined, scatter in data to be assessed and allows for the rapid identification of special or unusual features. A well laid out graph containing all the components discussed in this chapter can act as a 'one stop' summary of a whole ...
Abstract. This chapter covers the topics of data collection, data presentation and data analysis. It gives attention to data collection for studies based on experiments, on data derived from existing published or unpublished data sets, on observation, on simulation and digital twins, on surveys, on interviews and on focus group discussions.
Presenting Data - Graphs and Tables Types of Data. There are different types of data that can be collected in an experiment. Typically, we try to design experiments that collect objective, quantitative data. Objective data is fact-based, measurable, and observable. This means that if two people made the same measurement with the same tool ...
The way in which the results of experimental measurements can be best used in the extraction of conclusions relating to the magnitude measured is presented. The understanding of the concepts and methods presented in this chapter possibly constitute the main benefit the reader may derive from studying this book.
In one sample comparisons, the data of the experimental sample are compared to a specific reference value. The example in Table 1 is a comparison between the arterial pressures of the experimental sample with the reference value of 60 mmHg. Based on the distribution of experimental data, a parametric or non-parametric method of statistical ...
The Presentation of Data When significant amounts of quantitative data are presented in a report or publication, it is most effective to use tables and/or graphs. ... experimental results. The use of the broken line method mentioned above is often convenient in such a situation. 6. Too much information should not be included on ...
chapter, data is interpreted in a descriptive form. This chapter comprises the analysis, presentation and interpretation of the findings resulting from this study. The analysis and interpretation of data is carried out in two phases. The first part, which is based on the results of the questionnaire, deals with a quantitative analysis of data.
Appendix: Experimental Data and Plots This appendix presents the data, calculations, and graphs from the experiment to verify the ideal gas equation. The first two columns of Table A-1 show the measured voltages from the pressure transducer and the temperature transducer.
Effective data management is crucial for scientific integrity and reproducibility, a cornerstone of scientific progress. Well-organized and well-documented data enable validation and building on results. Data management encompasses activities including organization, documentation, storage, sharing, …
The GUI offers the procedures for data imputation, model establishment, model optimization and results presentation. Multivariate regression is a fundamental supervised chemometric method for developing the relationship between the independent variables and quantitative response, and it has been widely applied for data analysis in many research ...
In conjunction with historical losses, an estimated 35% of global mangrove area has been lost over the past five decades to human-driven land-use change, extreme weather events, and erosion (1-3).However, growing awareness around mangrove-dependent socio-ecological well-being has led to important conservation and restoration efforts of these ecosystems, with annual deforestation rates ...
To investigate the anchorage loss mechanism of externally prestressed CFRP tendons in concrete beams, this study introduces a novel theoretical calculation system (TCS) developed through both the finite element method (FEM) and experimental validation. Firstly, the FEM and the proposed TCS were employed based on the mechanism of anchorage loss to compute the deformation of each part of the ...
Analysis and Presentation of Experimental Results. 2 Report Writing. Reports may vary considerably in form depending upon the circumstances of the writer. The researcher working in a university or college will publish his findings in order to inform and, hopefully, gain the approbation of colleagues with similar interests.
Our experimental results underscore the superior performance of EduCross, demonstrating its effectiveness through the use of the real Multimodal Lecture Presentations dataset that mirrors authentic educational settings. These outcomes highlight the significant advancements of EduCross over existing methods, marking a leap forward in the ...
The presentation of physical quantities with their inaccuracies. 3. Errors: classification and propagation. 4. Probability distributions. 5. ... 5 - Processing of experimental data. Published online by Cambridge University Press: 05 June 2012 Herman J. C. Berendsen. Show author details Herman J. C. Berendsen
Ruthe Huang, ScM '19, works for Practical Intelligence as a Senior Data Scientist, where she specializes in statistical modeling and data visualization. Her thesis, under the direction of Mei-Cheng Wang, was entitled "ADHD Risk Prediction in the Boston Birth Cohort: an Application of the Proportional Odds Model and ROC Curve Analysis.". In her spare time, Huang volunteers for the Prison-to ...
Home Bancorp, Inc. 2024 Q2 - Results - Earnings Call Presentation. Jul. 18, 2024 12:59 PM ET Home Bancorp, Inc. (HBCP) Stock. ... Intel AI, data center executive set to take reins at Cornelis: report.