World-Class
Data Science
Learn with instructors from:

t-test Calculator
Table of contents
Welcome to our t-test calculator! Here you can not only easily perform one-sample t-tests , but also two-sample t-tests , as well as paired t-tests .
Do you prefer to find the p-value from t-test, or would you rather find the t-test critical values? Well, this t-test calculator can do both! 😊
What does a t-test tell you? Take a look at the text below, where we explain what actually gets tested when various types of t-tests are performed. Also, we explain when to use t-tests (in particular, whether to use the z-test vs. t-test) and what assumptions your data should satisfy for the results of a t-test to be valid. If you've ever wanted to know how to do a t-test by hand, we provide the necessary t-test formula, as well as tell you how to determine the number of degrees of freedom in a t-test.
When to use a t-test?
A t-test is one of the most popular statistical tests for location , i.e., it deals with the population(s) mean value(s).
There are different types of t-tests that you can perform:
- A one-sample t-test;
- A two-sample t-test; and
- A paired t-test.
In the next section , we explain when to use which. Remember that a t-test can only be used for one or two groups . If you need to compare three (or more) means, use the analysis of variance ( ANOVA ) method.
The t-test is a parametric test, meaning that your data has to fulfill some assumptions :
- The data points are independent; AND
- The data, at least approximately, follow a normal distribution .
If your sample doesn't fit these assumptions, you can resort to nonparametric alternatives. Visit our Mann–Whitney U test calculator or the Wilcoxon rank-sum test calculator to learn more. Other possibilities include the Wilcoxon signed-rank test or the sign test.
Which t-test?
Your choice of t-test depends on whether you are studying one group or two groups:
One sample t-test
Choose the one-sample t-test to check if the mean of a population is equal to some pre-set hypothesized value .
The average volume of a drink sold in 0.33 l cans — is it really equal to 330 ml?
The average weight of people from a specific city — is it different from the national average?
Two-sample t-test
Choose the two-sample t-test to check if the difference between the means of two populations is equal to some pre-determined value when the two samples have been chosen independently of each other.
In particular, you can use this test to check whether the two groups are different from one another .
The average difference in weight gain in two groups of people: one group was on a high-carb diet and the other on a high-fat diet.
The average difference in the results of a math test from students at two different universities.
This test is sometimes referred to as an independent samples t-test , or an unpaired samples t-test .
Paired t-test
A paired t-test is used to investigate the change in the mean of a population before and after some experimental intervention , based on a paired sample, i.e., when each subject has been measured twice: before and after treatment.
In particular, you can use this test to check whether, on average, the treatment has had any effect on the population .
The change in student test performance before and after taking a course.
The change in blood pressure in patients before and after administering some drug.
How to do a t-test?
So, you've decided which t-test to perform. These next steps will tell you how to calculate the p-value from t-test or its critical values, and then which decision to make about the null hypothesis.
Decide on the alternative hypothesis :
Use a two-tailed t-test if you only care whether the population's mean (or, in the case of two populations, the difference between the populations' means) agrees or disagrees with the pre-set value.
Use a one-tailed t-test if you want to test whether this mean (or difference in means) is greater/less than the pre-set value.
Compute your T-score value :
Formulas for the test statistic in t-tests include the sample size , as well as its mean and standard deviation . The exact formula depends on the t-test type — check the sections dedicated to each particular test for more details.
Determine the degrees of freedom for the t-test:
The degrees of freedom are the number of observations in a sample that are free to vary as we estimate statistical parameters. In the simplest case, the number of degrees of freedom equals your sample size minus the number of parameters you need to estimate . Again, the exact formula depends on the t-test you want to perform — check the sections below for details.
The degrees of freedom are essential, as they determine the distribution followed by your T-score (under the null hypothesis). If there are d degrees of freedom, then the distribution of the test statistics is the t-Student distribution with d degrees of freedom . This distribution has a shape similar to N(0,1) (bell-shaped and symmetric) but has heavier tails . If the number of degrees of freedom is large (>30), which generically happens for large samples, the t-Student distribution is practically indistinguishable from N(0,1).
💡 The t-Student distribution owes its name to William Sealy Gosset, who, in 1908, published his paper on the t-test under the pseudonym "Student". Gosset worked at the famous Guinness Brewery in Dublin, Ireland, and devised the t-test as an economical way to monitor the quality of beer. Cheers! 🍺🍺🍺
p-value from t-test
Recall that the p-value is the probability (calculated under the assumption that the null hypothesis is true) that the test statistic will produce values at least as extreme as the T-score produced for your sample . As probabilities correspond to areas under the density function, p-value from t-test can be nicely illustrated with the help of the following pictures:
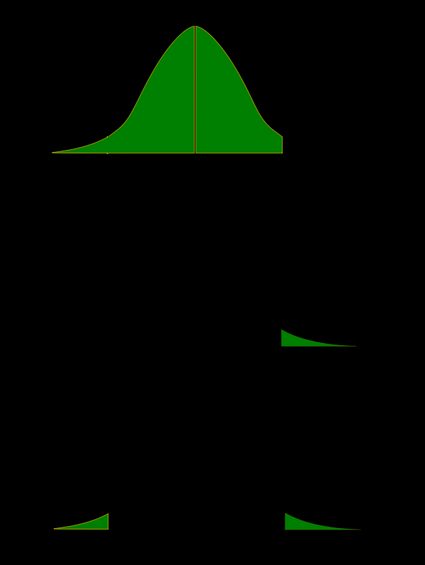
The following formulae say how to calculate p-value from t-test. By cdf t,d we denote the cumulative distribution function of the t-Student distribution with d degrees of freedom:
p-value from left-tailed t-test:
p-value = cdf t,d (t score )
p-value from right-tailed t-test:
p-value = 1 − cdf t,d (t score )
p-value from two-tailed t-test:
p-value = 2 × cdf t,d (−|t score |)
or, equivalently: p-value = 2 − 2 × cdf t,d (|t score |)
However, the cdf of the t-distribution is given by a somewhat complicated formula. To find the p-value by hand, you would need to resort to statistical tables, where approximate cdf values are collected, or to specialized statistical software. Fortunately, our t-test calculator determines the p-value from t-test for you in the blink of an eye!
t-test critical values
Recall, that in the critical values approach to hypothesis testing, you need to set a significance level, α, before computing the critical values , which in turn give rise to critical regions (a.k.a. rejection regions).
Formulas for critical values employ the quantile function of t-distribution, i.e., the inverse of the cdf :
Critical value for left-tailed t-test: cdf t,d -1 (α)
critical region:
(-∞, cdf t,d -1 (α)]
Critical value for right-tailed t-test: cdf t,d -1 (1-α)
[cdf t,d -1 (1-α), ∞)
Critical values for two-tailed t-test: ±cdf t,d -1 (1-α/2)
(-∞, -cdf t,d -1 (1-α/2)] ∪ [cdf t,d -1 (1-α/2), ∞)
To decide the fate of the null hypothesis, just check if your T-score lies within the critical region:
If your T-score belongs to the critical region , reject the null hypothesis and accept the alternative hypothesis.
If your T-score is outside the critical region , then you don't have enough evidence to reject the null hypothesis.
How to use our t-test calculator
Choose the type of t-test you wish to perform:
A one-sample t-test (to test the mean of a single group against a hypothesized mean);
A two-sample t-test (to compare the means for two groups); or
A paired t-test (to check how the mean from the same group changes after some intervention).
Two-tailed;
Left-tailed; or
Right-tailed.
This t-test calculator allows you to use either the p-value approach or the critical regions approach to hypothesis testing!
Enter your T-score and the number of degrees of freedom . If you don't know them, provide some data about your sample(s): sample size, mean, and standard deviation, and our t-test calculator will compute the T-score and degrees of freedom for you .
Once all the parameters are present, the p-value, or critical region, will immediately appear underneath the t-test calculator, along with an interpretation!
One-sample t-test
The null hypothesis is that the population mean is equal to some value μ 0 \mu_0 μ 0 .
The alternative hypothesis is that the population mean is:
- different from μ 0 \mu_0 μ 0 ;
- smaller than μ 0 \mu_0 μ 0 ; or
- greater than μ 0 \mu_0 μ 0 .
One-sample t-test formula :
- μ 0 \mu_0 μ 0 — Mean postulated in the null hypothesis;
- n n n — Sample size;
- x ˉ \bar{x} x ˉ — Sample mean; and
- s s s — Sample standard deviation.
Number of degrees of freedom in t-test (one-sample) = n − 1 n-1 n − 1 .
The null hypothesis is that the actual difference between these groups' means, μ 1 \mu_1 μ 1 , and μ 2 \mu_2 μ 2 , is equal to some pre-set value, Δ \Delta Δ .
The alternative hypothesis is that the difference μ 1 − μ 2 \mu_1 - \mu_2 μ 1 − μ 2 is:
- Different from Δ \Delta Δ ;
- Smaller than Δ \Delta Δ ; or
- Greater than Δ \Delta Δ .
In particular, if this pre-determined difference is zero ( Δ = 0 \Delta = 0 Δ = 0 ):
The null hypothesis is that the population means are equal.
The alternate hypothesis is that the population means are:
- μ 1 \mu_1 μ 1 and μ 2 \mu_2 μ 2 are different from one another;
- μ 1 \mu_1 μ 1 is smaller than μ 2 \mu_2 μ 2 ; and
- μ 1 \mu_1 μ 1 is greater than μ 2 \mu_2 μ 2 .
Formally, to perform a t-test, we should additionally assume that the variances of the two populations are equal (this assumption is called the homogeneity of variance ).
There is a version of a t-test that can be applied without the assumption of homogeneity of variance: it is called a Welch's t-test . For your convenience, we describe both versions.
Two-sample t-test if variances are equal
Use this test if you know that the two populations' variances are the same (or very similar).
Two-sample t-test formula (with equal variances) :
where s p s_p s p is the so-called pooled standard deviation , which we compute as:
- Δ \Delta Δ — Mean difference postulated in the null hypothesis;
- n 1 n_1 n 1 — First sample size;
- x ˉ 1 \bar{x}_1 x ˉ 1 — Mean for the first sample;
- s 1 s_1 s 1 — Standard deviation in the first sample;
- n 2 n_2 n 2 — Second sample size;
- x ˉ 2 \bar{x}_2 x ˉ 2 — Mean for the second sample; and
- s 2 s_2 s 2 — Standard deviation in the second sample.
Number of degrees of freedom in t-test (two samples, equal variances) = n 1 + n 2 − 2 n_1 + n_2 - 2 n 1 + n 2 − 2 .
Two-sample t-test if variances are unequal (Welch's t-test)
Use this test if the variances of your populations are different.
Two-sample Welch's t-test formula if variances are unequal:
- s 1 s_1 s 1 — Standard deviation in the first sample;
- s 2 s_2 s 2 — Standard deviation in the second sample.
The number of degrees of freedom in a Welch's t-test (two-sample t-test with unequal variances) is very difficult to count. We can approximate it with the help of the following Satterthwaite formula :
Alternatively, you can take the smaller of n 1 − 1 n_1 - 1 n 1 − 1 and n 2 − 1 n_2 - 1 n 2 − 1 as a conservative estimate for the number of degrees of freedom.
🔎 The Satterthwaite formula for the degrees of freedom can be rewritten as a scaled weighted harmonic mean of the degrees of freedom of the respective samples: n 1 − 1 n_1 - 1 n 1 − 1 and n 2 − 1 n_2 - 1 n 2 − 1 , and the weights are proportional to the standard deviations of the corresponding samples.
As we commonly perform a paired t-test when we have data about the same subjects measured twice (before and after some treatment), let us adopt the convention of referring to the samples as the pre-group and post-group.
The null hypothesis is that the true difference between the means of pre- and post-populations is equal to some pre-set value, Δ \Delta Δ .
The alternative hypothesis is that the actual difference between these means is:
Typically, this pre-determined difference is zero. We can then reformulate the hypotheses as follows:
The null hypothesis is that the pre- and post-means are the same, i.e., the treatment has no impact on the population .
The alternative hypothesis:
- The pre- and post-means are different from one another (treatment has some effect);
- The pre-mean is smaller than the post-mean (treatment increases the result); or
- The pre-mean is greater than the post-mean (treatment decreases the result).
Paired t-test formula
In fact, a paired t-test is technically the same as a one-sample t-test! Let us see why it is so. Let x 1 , . . . , x n x_1, ... , x_n x 1 , ... , x n be the pre observations and y 1 , . . . , y n y_1, ... , y_n y 1 , ... , y n the respective post observations. That is, x i , y i x_i, y_i x i , y i are the before and after measurements of the i -th subject.
For each subject, compute the difference, d i : = x i − y i d_i := x_i - y_i d i := x i − y i . All that happens next is just a one-sample t-test performed on the sample of differences d 1 , . . . , d n d_1, ... , d_n d 1 , ... , d n . Take a look at the formula for the T-score :
Δ \Delta Δ — Mean difference postulated in the null hypothesis;
n n n — Size of the sample of differences, i.e., the number of pairs;
x ˉ \bar{x} x ˉ — Mean of the sample of differences; and
s s s — Standard deviation of the sample of differences.
Number of degrees of freedom in t-test (paired): n − 1 n - 1 n − 1
t-test vs Z-test
We use a Z-test when we want to test the population mean of a normally distributed dataset, which has a known population variance . If the number of degrees of freedom is large, then the t-Student distribution is very close to N(0,1).
Hence, if there are many data points (at least 30), you may swap a t-test for a Z-test, and the results will be almost identical. However, for small samples with unknown variance, remember to use the t-test because, in such cases, the t-Student distribution differs significantly from the N(0,1)!
🙋 Have you concluded you need to perform the z-test? Head straight to our z-test calculator !
What is a t-test?
A t-test is a widely used statistical test that analyzes the means of one or two groups of data. For instance, a t-test is performed on medical data to determine whether a new drug really helps.
What are different types of t-tests?
Different types of t-tests are:
- One-sample t-test;
- Two-sample t-test; and
- Paired t-test.
How to find the t value in a one sample t-test?
To find the t-value:
- Subtract the null hypothesis mean from the sample mean value.
- Divide the difference by the standard deviation of the sample.
- Multiply the resultant with the square root of the sample size.
.css-m482sy.css-m482sy{color:#2B3148;background-color:transparent;font-family:var(--calculator-ui-font-family),Verdana,sans-serif;font-size:20px;line-height:24px;overflow:visible;padding-top:0px;position:relative;}.css-m482sy.css-m482sy:after{content:'';-webkit-transform:scale(0);-moz-transform:scale(0);-ms-transform:scale(0);transform:scale(0);position:absolute;border:2px solid #EA9430;border-radius:2px;inset:-8px;z-index:1;}.css-m482sy .js-external-link-button.link-like,.css-m482sy .js-external-link-anchor{color:inherit;border-radius:1px;-webkit-text-decoration:underline;text-decoration:underline;}.css-m482sy .js-external-link-button.link-like:hover,.css-m482sy .js-external-link-anchor:hover,.css-m482sy .js-external-link-button.link-like:active,.css-m482sy .js-external-link-anchor:active{text-decoration-thickness:2px;text-shadow:1px 0 0;}.css-m482sy .js-external-link-button.link-like:focus-visible,.css-m482sy .js-external-link-anchor:focus-visible{outline:transparent 2px dotted;box-shadow:0 0 0 2px #6314E6;}.css-m482sy p,.css-m482sy div{margin:0;display:block;}.css-m482sy pre{margin:0;display:block;}.css-m482sy pre code{display:block;width:-webkit-fit-content;width:-moz-fit-content;width:fit-content;}.css-m482sy pre:not(:first-child){padding-top:8px;}.css-m482sy ul,.css-m482sy ol{display:block margin:0;padding-left:20px;}.css-m482sy ul li,.css-m482sy ol li{padding-top:8px;}.css-m482sy ul ul,.css-m482sy ol ul,.css-m482sy ul ol,.css-m482sy ol ol{padding-top:0;}.css-m482sy ul:not(:first-child),.css-m482sy ol:not(:first-child){padding-top:4px;} .css-63uqft{margin:auto;background-color:white;overflow:auto;overflow-wrap:break-word;word-break:break-word;}.css-63uqft code,.css-63uqft kbd,.css-63uqft pre,.css-63uqft samp{font-family:monospace;}.css-63uqft code{padding:2px 4px;color:#444;background:#ddd;border-radius:4px;}.css-63uqft figcaption,.css-63uqft caption{text-align:center;}.css-63uqft figcaption{font-size:12px;font-style:italic;overflow:hidden;}.css-63uqft h3{font-size:1.75rem;}.css-63uqft h4{font-size:1.5rem;}.css-63uqft .mathBlock{font-size:24px;-webkit-padding-start:4px;padding-inline-start:4px;}.css-63uqft .mathBlock .katex{font-size:24px;text-align:left;}.css-63uqft .math-inline{background-color:#f0f0f0;display:inline-block;font-size:inherit;padding:0 3px;}.css-63uqft .videoBlock,.css-63uqft .imageBlock{margin-bottom:16px;}.css-63uqft .imageBlock__image-align--left,.css-63uqft .videoBlock__video-align--left{float:left;}.css-63uqft .imageBlock__image-align--right,.css-63uqft .videoBlock__video-align--right{float:right;}.css-63uqft .imageBlock__image-align--center,.css-63uqft .videoBlock__video-align--center{display:block;margin-left:auto;margin-right:auto;clear:both;}.css-63uqft .imageBlock__image-align--none,.css-63uqft .videoBlock__video-align--none{clear:both;margin-left:0;margin-right:0;}.css-63uqft .videoBlock__video--wrapper{position:relative;padding-bottom:56.25%;height:0;}.css-63uqft .videoBlock__video--wrapper iframe{position:absolute;top:0;left:0;width:100%;height:100%;}.css-63uqft .videoBlock__caption{text-align:left;}@font-face{font-family:'KaTeX_AMS';src:url(/katex-fonts/KaTeX_AMS-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_AMS-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_AMS-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Caligraphic';src:url(/katex-fonts/KaTeX_Caligraphic-Bold.woff2) format('woff2'),url(/katex-fonts/KaTeX_Caligraphic-Bold.woff) format('woff'),url(/katex-fonts/KaTeX_Caligraphic-Bold.ttf) format('truetype');font-weight:bold;font-style:normal;}@font-face{font-family:'KaTeX_Caligraphic';src:url(/katex-fonts/KaTeX_Caligraphic-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Caligraphic-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Caligraphic-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Fraktur';src:url(/katex-fonts/KaTeX_Fraktur-Bold.woff2) format('woff2'),url(/katex-fonts/KaTeX_Fraktur-Bold.woff) format('woff'),url(/katex-fonts/KaTeX_Fraktur-Bold.ttf) format('truetype');font-weight:bold;font-style:normal;}@font-face{font-family:'KaTeX_Fraktur';src:url(/katex-fonts/KaTeX_Fraktur-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Fraktur-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Fraktur-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Main';src:url(/katex-fonts/KaTeX_Main-Bold.woff2) format('woff2'),url(/katex-fonts/KaTeX_Main-Bold.woff) format('woff'),url(/katex-fonts/KaTeX_Main-Bold.ttf) format('truetype');font-weight:bold;font-style:normal;}@font-face{font-family:'KaTeX_Main';src:url(/katex-fonts/KaTeX_Main-BoldItalic.woff2) format('woff2'),url(/katex-fonts/KaTeX_Main-BoldItalic.woff) format('woff'),url(/katex-fonts/KaTeX_Main-BoldItalic.ttf) format('truetype');font-weight:bold;font-style:italic;}@font-face{font-family:'KaTeX_Main';src:url(/katex-fonts/KaTeX_Main-Italic.woff2) format('woff2'),url(/katex-fonts/KaTeX_Main-Italic.woff) format('woff'),url(/katex-fonts/KaTeX_Main-Italic.ttf) format('truetype');font-weight:normal;font-style:italic;}@font-face{font-family:'KaTeX_Main';src:url(/katex-fonts/KaTeX_Main-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Main-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Main-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Math';src:url(/katex-fonts/KaTeX_Math-BoldItalic.woff2) format('woff2'),url(/katex-fonts/KaTeX_Math-BoldItalic.woff) format('woff'),url(/katex-fonts/KaTeX_Math-BoldItalic.ttf) format('truetype');font-weight:bold;font-style:italic;}@font-face{font-family:'KaTeX_Math';src:url(/katex-fonts/KaTeX_Math-Italic.woff2) format('woff2'),url(/katex-fonts/KaTeX_Math-Italic.woff) format('woff'),url(/katex-fonts/KaTeX_Math-Italic.ttf) format('truetype');font-weight:normal;font-style:italic;}@font-face{font-family:'KaTeX_SansSerif';src:url(/katex-fonts/KaTeX_SansSerif-Bold.woff2) format('woff2'),url(/katex-fonts/KaTeX_SansSerif-Bold.woff) format('woff'),url(/katex-fonts/KaTeX_SansSerif-Bold.ttf) format('truetype');font-weight:bold;font-style:normal;}@font-face{font-family:'KaTeX_SansSerif';src:url(/katex-fonts/KaTeX_SansSerif-Italic.woff2) format('woff2'),url(/katex-fonts/KaTeX_SansSerif-Italic.woff) format('woff'),url(/katex-fonts/KaTeX_SansSerif-Italic.ttf) format('truetype');font-weight:normal;font-style:italic;}@font-face{font-family:'KaTeX_SansSerif';src:url(/katex-fonts/KaTeX_SansSerif-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_SansSerif-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_SansSerif-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Script';src:url(/katex-fonts/KaTeX_Script-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Script-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Script-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Size1';src:url(/katex-fonts/KaTeX_Size1-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Size1-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Size1-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Size2';src:url(/katex-fonts/KaTeX_Size2-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Size2-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Size2-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Size3';src:url(/katex-fonts/KaTeX_Size3-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Size3-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Size3-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Size4';src:url(/katex-fonts/KaTeX_Size4-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Size4-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Size4-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}@font-face{font-family:'KaTeX_Typewriter';src:url(/katex-fonts/KaTeX_Typewriter-Regular.woff2) format('woff2'),url(/katex-fonts/KaTeX_Typewriter-Regular.woff) format('woff'),url(/katex-fonts/KaTeX_Typewriter-Regular.ttf) format('truetype');font-weight:normal;font-style:normal;}.css-63uqft .katex{font:normal 1.21em KaTeX_Main,Times New Roman,serif;line-height:1.2;text-indent:0;text-rendering:auto;}.css-63uqft .katex *{-ms-high-contrast-adjust:none!important;border-color:currentColor;}.css-63uqft .katex .katex-version::after{content:'0.13.13';}.css-63uqft .katex .katex-mathml{position:absolute;clip:rect(1px,1px,1px,1px);padding:0;border:0;height:1px;width:1px;overflow:hidden;}.css-63uqft .katex .katex-html>.newline{display:block;}.css-63uqft .katex .base{position:relative;display:inline-block;white-space:nowrap;width:-webkit-min-content;width:-moz-min-content;width:-webkit-min-content;width:-moz-min-content;width:min-content;}.css-63uqft .katex .strut{display:inline-block;}.css-63uqft .katex .textbf{font-weight:bold;}.css-63uqft .katex .textit{font-style:italic;}.css-63uqft .katex .textrm{font-family:KaTeX_Main;}.css-63uqft .katex .textsf{font-family:KaTeX_SansSerif;}.css-63uqft .katex .texttt{font-family:KaTeX_Typewriter;}.css-63uqft .katex .mathnormal{font-family:KaTeX_Math;font-style:italic;}.css-63uqft .katex .mathit{font-family:KaTeX_Main;font-style:italic;}.css-63uqft .katex .mathrm{font-style:normal;}.css-63uqft .katex .mathbf{font-family:KaTeX_Main;font-weight:bold;}.css-63uqft .katex .boldsymbol{font-family:KaTeX_Math;font-weight:bold;font-style:italic;}.css-63uqft .katex .amsrm{font-family:KaTeX_AMS;}.css-63uqft .katex .mathbb,.css-63uqft .katex .textbb{font-family:KaTeX_AMS;}.css-63uqft .katex .mathcal{font-family:KaTeX_Caligraphic;}.css-63uqft .katex .mathfrak,.css-63uqft .katex .textfrak{font-family:KaTeX_Fraktur;}.css-63uqft .katex .mathtt{font-family:KaTeX_Typewriter;}.css-63uqft .katex .mathscr,.css-63uqft .katex .textscr{font-family:KaTeX_Script;}.css-63uqft .katex .mathsf,.css-63uqft .katex .textsf{font-family:KaTeX_SansSerif;}.css-63uqft .katex .mathboldsf,.css-63uqft .katex .textboldsf{font-family:KaTeX_SansSerif;font-weight:bold;}.css-63uqft .katex .mathitsf,.css-63uqft .katex .textitsf{font-family:KaTeX_SansSerif;font-style:italic;}.css-63uqft .katex .mainrm{font-family:KaTeX_Main;font-style:normal;}.css-63uqft .katex .vlist-t{display:inline-table;table-layout:fixed;border-collapse:collapse;}.css-63uqft .katex .vlist-r{display:table-row;}.css-63uqft .katex .vlist{display:table-cell;vertical-align:bottom;position:relative;}.css-63uqft .katex .vlist>span{display:block;height:0;position:relative;}.css-63uqft .katex .vlist>span>span{display:inline-block;}.css-63uqft .katex .vlist>span>.pstrut{overflow:hidden;width:0;}.css-63uqft .katex .vlist-t2{margin-right:-2px;}.css-63uqft .katex .vlist-s{display:table-cell;vertical-align:bottom;font-size:1px;width:2px;min-width:2px;}.css-63uqft .katex .vbox{display:-webkit-inline-box;display:-webkit-inline-flex;display:-ms-inline-flexbox;display:inline-flex;-webkit-flex-direction:column;-ms-flex-direction:column;flex-direction:column;-webkit-align-items:baseline;-webkit-box-align:baseline;-ms-flex-align:baseline;align-items:baseline;}.css-63uqft .katex .hbox{display:-webkit-inline-box;display:-webkit-inline-flex;display:-ms-inline-flexbox;display:inline-flex;-webkit-flex-direction:row;-ms-flex-direction:row;flex-direction:row;width:100%;}.css-63uqft .katex .thinbox{display:-webkit-inline-box;display:-webkit-inline-flex;display:-ms-inline-flexbox;display:inline-flex;-webkit-flex-direction:row;-ms-flex-direction:row;flex-direction:row;width:0;max-width:0;}.css-63uqft .katex .msupsub{text-align:left;}.css-63uqft .katex .mfrac>span>span{text-align:center;}.css-63uqft .katex .mfrac .frac-line{display:inline-block;width:100%;border-bottom-style:solid;}.css-63uqft .katex .mfrac .frac-line,.css-63uqft .katex .overline .overline-line,.css-63uqft .katex .underline .underline-line,.css-63uqft .katex .hline,.css-63uqft .katex .hdashline,.css-63uqft .katex .rule{min-height:1px;}.css-63uqft .katex .mspace{display:inline-block;}.css-63uqft .katex .llap,.css-63uqft .katex .rlap,.css-63uqft .katex .clap{width:0;position:relative;}.css-63uqft .katex .llap>.inner,.css-63uqft .katex .rlap>.inner,.css-63uqft .katex .clap>.inner{position:absolute;}.css-63uqft .katex .llap>.fix,.css-63uqft .katex .rlap>.fix,.css-63uqft .katex .clap>.fix{display:inline-block;}.css-63uqft .katex .llap>.inner{right:0;}.css-63uqft .katex .rlap>.inner,.css-63uqft .katex .clap>.inner{left:0;}.css-63uqft .katex .clap>.inner>span{margin-left:-50%;margin-right:50%;}.css-63uqft .katex .rule{display:inline-block;border:solid 0;position:relative;}.css-63uqft .katex .overline .overline-line,.css-63uqft .katex .underline .underline-line,.css-63uqft .katex .hline{display:inline-block;width:100%;border-bottom-style:solid;}.css-63uqft .katex .hdashline{display:inline-block;width:100%;border-bottom-style:dashed;}.css-63uqft .katex .sqrt>.root{margin-left:0.27777778em;margin-right:-0.55555556em;}.css-63uqft .katex .sizing.reset-size1.size1,.css-63uqft .katex .fontsize-ensurer.reset-size1.size1{font-size:1em;}.css-63uqft .katex .sizing.reset-size1.size2,.css-63uqft .katex .fontsize-ensurer.reset-size1.size2{font-size:1.2em;}.css-63uqft .katex .sizing.reset-size1.size3,.css-63uqft .katex .fontsize-ensurer.reset-size1.size3{font-size:1.4em;}.css-63uqft .katex .sizing.reset-size1.size4,.css-63uqft .katex .fontsize-ensurer.reset-size1.size4{font-size:1.6em;}.css-63uqft .katex .sizing.reset-size1.size5,.css-63uqft .katex .fontsize-ensurer.reset-size1.size5{font-size:1.8em;}.css-63uqft .katex .sizing.reset-size1.size6,.css-63uqft .katex .fontsize-ensurer.reset-size1.size6{font-size:2em;}.css-63uqft .katex .sizing.reset-size1.size7,.css-63uqft .katex .fontsize-ensurer.reset-size1.size7{font-size:2.4em;}.css-63uqft .katex .sizing.reset-size1.size8,.css-63uqft .katex .fontsize-ensurer.reset-size1.size8{font-size:2.88em;}.css-63uqft .katex .sizing.reset-size1.size9,.css-63uqft .katex .fontsize-ensurer.reset-size1.size9{font-size:3.456em;}.css-63uqft .katex .sizing.reset-size1.size10,.css-63uqft .katex .fontsize-ensurer.reset-size1.size10{font-size:4.148em;}.css-63uqft .katex .sizing.reset-size1.size11,.css-63uqft .katex .fontsize-ensurer.reset-size1.size11{font-size:4.976em;}.css-63uqft .katex .sizing.reset-size2.size1,.css-63uqft .katex .fontsize-ensurer.reset-size2.size1{font-size:0.83333333em;}.css-63uqft .katex .sizing.reset-size2.size2,.css-63uqft .katex .fontsize-ensurer.reset-size2.size2{font-size:1em;}.css-63uqft .katex .sizing.reset-size2.size3,.css-63uqft .katex .fontsize-ensurer.reset-size2.size3{font-size:1.16666667em;}.css-63uqft .katex .sizing.reset-size2.size4,.css-63uqft .katex .fontsize-ensurer.reset-size2.size4{font-size:1.33333333em;}.css-63uqft .katex .sizing.reset-size2.size5,.css-63uqft .katex .fontsize-ensurer.reset-size2.size5{font-size:1.5em;}.css-63uqft .katex .sizing.reset-size2.size6,.css-63uqft .katex .fontsize-ensurer.reset-size2.size6{font-size:1.66666667em;}.css-63uqft .katex .sizing.reset-size2.size7,.css-63uqft .katex .fontsize-ensurer.reset-size2.size7{font-size:2em;}.css-63uqft .katex .sizing.reset-size2.size8,.css-63uqft .katex .fontsize-ensurer.reset-size2.size8{font-size:2.4em;}.css-63uqft .katex .sizing.reset-size2.size9,.css-63uqft .katex .fontsize-ensurer.reset-size2.size9{font-size:2.88em;}.css-63uqft .katex .sizing.reset-size2.size10,.css-63uqft .katex .fontsize-ensurer.reset-size2.size10{font-size:3.45666667em;}.css-63uqft .katex .sizing.reset-size2.size11,.css-63uqft .katex .fontsize-ensurer.reset-size2.size11{font-size:4.14666667em;}.css-63uqft .katex .sizing.reset-size3.size1,.css-63uqft .katex .fontsize-ensurer.reset-size3.size1{font-size:0.71428571em;}.css-63uqft .katex .sizing.reset-size3.size2,.css-63uqft .katex .fontsize-ensurer.reset-size3.size2{font-size:0.85714286em;}.css-63uqft .katex .sizing.reset-size3.size3,.css-63uqft .katex .fontsize-ensurer.reset-size3.size3{font-size:1em;}.css-63uqft .katex .sizing.reset-size3.size4,.css-63uqft .katex .fontsize-ensurer.reset-size3.size4{font-size:1.14285714em;}.css-63uqft .katex .sizing.reset-size3.size5,.css-63uqft .katex .fontsize-ensurer.reset-size3.size5{font-size:1.28571429em;}.css-63uqft .katex .sizing.reset-size3.size6,.css-63uqft .katex .fontsize-ensurer.reset-size3.size6{font-size:1.42857143em;}.css-63uqft .katex .sizing.reset-size3.size7,.css-63uqft .katex .fontsize-ensurer.reset-size3.size7{font-size:1.71428571em;}.css-63uqft .katex .sizing.reset-size3.size8,.css-63uqft .katex .fontsize-ensurer.reset-size3.size8{font-size:2.05714286em;}.css-63uqft .katex .sizing.reset-size3.size9,.css-63uqft .katex .fontsize-ensurer.reset-size3.size9{font-size:2.46857143em;}.css-63uqft .katex .sizing.reset-size3.size10,.css-63uqft .katex .fontsize-ensurer.reset-size3.size10{font-size:2.96285714em;}.css-63uqft .katex .sizing.reset-size3.size11,.css-63uqft .katex .fontsize-ensurer.reset-size3.size11{font-size:3.55428571em;}.css-63uqft .katex .sizing.reset-size4.size1,.css-63uqft .katex .fontsize-ensurer.reset-size4.size1{font-size:0.625em;}.css-63uqft .katex .sizing.reset-size4.size2,.css-63uqft .katex .fontsize-ensurer.reset-size4.size2{font-size:0.75em;}.css-63uqft .katex .sizing.reset-size4.size3,.css-63uqft .katex .fontsize-ensurer.reset-size4.size3{font-size:0.875em;}.css-63uqft .katex .sizing.reset-size4.size4,.css-63uqft .katex .fontsize-ensurer.reset-size4.size4{font-size:1em;}.css-63uqft .katex .sizing.reset-size4.size5,.css-63uqft .katex .fontsize-ensurer.reset-size4.size5{font-size:1.125em;}.css-63uqft .katex .sizing.reset-size4.size6,.css-63uqft .katex .fontsize-ensurer.reset-size4.size6{font-size:1.25em;}.css-63uqft .katex .sizing.reset-size4.size7,.css-63uqft .katex .fontsize-ensurer.reset-size4.size7{font-size:1.5em;}.css-63uqft .katex .sizing.reset-size4.size8,.css-63uqft .katex .fontsize-ensurer.reset-size4.size8{font-size:1.8em;}.css-63uqft .katex .sizing.reset-size4.size9,.css-63uqft .katex .fontsize-ensurer.reset-size4.size9{font-size:2.16em;}.css-63uqft .katex .sizing.reset-size4.size10,.css-63uqft .katex .fontsize-ensurer.reset-size4.size10{font-size:2.5925em;}.css-63uqft .katex .sizing.reset-size4.size11,.css-63uqft .katex .fontsize-ensurer.reset-size4.size11{font-size:3.11em;}.css-63uqft .katex .sizing.reset-size5.size1,.css-63uqft .katex .fontsize-ensurer.reset-size5.size1{font-size:0.55555556em;}.css-63uqft .katex .sizing.reset-size5.size2,.css-63uqft .katex .fontsize-ensurer.reset-size5.size2{font-size:0.66666667em;}.css-63uqft .katex .sizing.reset-size5.size3,.css-63uqft .katex .fontsize-ensurer.reset-size5.size3{font-size:0.77777778em;}.css-63uqft .katex .sizing.reset-size5.size4,.css-63uqft .katex .fontsize-ensurer.reset-size5.size4{font-size:0.88888889em;}.css-63uqft .katex .sizing.reset-size5.size5,.css-63uqft .katex .fontsize-ensurer.reset-size5.size5{font-size:1em;}.css-63uqft .katex .sizing.reset-size5.size6,.css-63uqft .katex .fontsize-ensurer.reset-size5.size6{font-size:1.11111111em;}.css-63uqft .katex .sizing.reset-size5.size7,.css-63uqft .katex .fontsize-ensurer.reset-size5.size7{font-size:1.33333333em;}.css-63uqft .katex .sizing.reset-size5.size8,.css-63uqft .katex .fontsize-ensurer.reset-size5.size8{font-size:1.6em;}.css-63uqft .katex .sizing.reset-size5.size9,.css-63uqft .katex .fontsize-ensurer.reset-size5.size9{font-size:1.92em;}.css-63uqft .katex .sizing.reset-size5.size10,.css-63uqft .katex .fontsize-ensurer.reset-size5.size10{font-size:2.30444444em;}.css-63uqft .katex .sizing.reset-size5.size11,.css-63uqft .katex .fontsize-ensurer.reset-size5.size11{font-size:2.76444444em;}.css-63uqft .katex .sizing.reset-size6.size1,.css-63uqft .katex .fontsize-ensurer.reset-size6.size1{font-size:0.5em;}.css-63uqft .katex .sizing.reset-size6.size2,.css-63uqft .katex .fontsize-ensurer.reset-size6.size2{font-size:0.6em;}.css-63uqft .katex .sizing.reset-size6.size3,.css-63uqft .katex .fontsize-ensurer.reset-size6.size3{font-size:0.7em;}.css-63uqft .katex .sizing.reset-size6.size4,.css-63uqft .katex .fontsize-ensurer.reset-size6.size4{font-size:0.8em;}.css-63uqft .katex .sizing.reset-size6.size5,.css-63uqft .katex .fontsize-ensurer.reset-size6.size5{font-size:0.9em;}.css-63uqft .katex .sizing.reset-size6.size6,.css-63uqft .katex .fontsize-ensurer.reset-size6.size6{font-size:1em;}.css-63uqft .katex .sizing.reset-size6.size7,.css-63uqft .katex .fontsize-ensurer.reset-size6.size7{font-size:1.2em;}.css-63uqft .katex .sizing.reset-size6.size8,.css-63uqft .katex .fontsize-ensurer.reset-size6.size8{font-size:1.44em;}.css-63uqft .katex .sizing.reset-size6.size9,.css-63uqft .katex .fontsize-ensurer.reset-size6.size9{font-size:1.728em;}.css-63uqft .katex .sizing.reset-size6.size10,.css-63uqft .katex .fontsize-ensurer.reset-size6.size10{font-size:2.074em;}.css-63uqft .katex .sizing.reset-size6.size11,.css-63uqft .katex .fontsize-ensurer.reset-size6.size11{font-size:2.488em;}.css-63uqft .katex .sizing.reset-size7.size1,.css-63uqft .katex .fontsize-ensurer.reset-size7.size1{font-size:0.41666667em;}.css-63uqft .katex .sizing.reset-size7.size2,.css-63uqft .katex .fontsize-ensurer.reset-size7.size2{font-size:0.5em;}.css-63uqft .katex .sizing.reset-size7.size3,.css-63uqft .katex .fontsize-ensurer.reset-size7.size3{font-size:0.58333333em;}.css-63uqft .katex .sizing.reset-size7.size4,.css-63uqft .katex .fontsize-ensurer.reset-size7.size4{font-size:0.66666667em;}.css-63uqft .katex .sizing.reset-size7.size5,.css-63uqft .katex .fontsize-ensurer.reset-size7.size5{font-size:0.75em;}.css-63uqft .katex .sizing.reset-size7.size6,.css-63uqft .katex .fontsize-ensurer.reset-size7.size6{font-size:0.83333333em;}.css-63uqft .katex .sizing.reset-size7.size7,.css-63uqft .katex .fontsize-ensurer.reset-size7.size7{font-size:1em;}.css-63uqft .katex .sizing.reset-size7.size8,.css-63uqft .katex .fontsize-ensurer.reset-size7.size8{font-size:1.2em;}.css-63uqft .katex .sizing.reset-size7.size9,.css-63uqft .katex .fontsize-ensurer.reset-size7.size9{font-size:1.44em;}.css-63uqft .katex .sizing.reset-size7.size10,.css-63uqft .katex .fontsize-ensurer.reset-size7.size10{font-size:1.72833333em;}.css-63uqft .katex .sizing.reset-size7.size11,.css-63uqft .katex .fontsize-ensurer.reset-size7.size11{font-size:2.07333333em;}.css-63uqft .katex .sizing.reset-size8.size1,.css-63uqft .katex .fontsize-ensurer.reset-size8.size1{font-size:0.34722222em;}.css-63uqft .katex .sizing.reset-size8.size2,.css-63uqft .katex .fontsize-ensurer.reset-size8.size2{font-size:0.41666667em;}.css-63uqft .katex .sizing.reset-size8.size3,.css-63uqft .katex .fontsize-ensurer.reset-size8.size3{font-size:0.48611111em;}.css-63uqft .katex .sizing.reset-size8.size4,.css-63uqft .katex .fontsize-ensurer.reset-size8.size4{font-size:0.55555556em;}.css-63uqft .katex .sizing.reset-size8.size5,.css-63uqft .katex .fontsize-ensurer.reset-size8.size5{font-size:0.625em;}.css-63uqft .katex .sizing.reset-size8.size6,.css-63uqft .katex .fontsize-ensurer.reset-size8.size6{font-size:0.69444444em;}.css-63uqft .katex .sizing.reset-size8.size7,.css-63uqft .katex .fontsize-ensurer.reset-size8.size7{font-size:0.83333333em;}.css-63uqft .katex .sizing.reset-size8.size8,.css-63uqft .katex .fontsize-ensurer.reset-size8.size8{font-size:1em;}.css-63uqft .katex .sizing.reset-size8.size9,.css-63uqft .katex .fontsize-ensurer.reset-size8.size9{font-size:1.2em;}.css-63uqft .katex .sizing.reset-size8.size10,.css-63uqft .katex .fontsize-ensurer.reset-size8.size10{font-size:1.44027778em;}.css-63uqft .katex .sizing.reset-size8.size11,.css-63uqft .katex .fontsize-ensurer.reset-size8.size11{font-size:1.72777778em;}.css-63uqft .katex .sizing.reset-size9.size1,.css-63uqft .katex .fontsize-ensurer.reset-size9.size1{font-size:0.28935185em;}.css-63uqft .katex .sizing.reset-size9.size2,.css-63uqft .katex .fontsize-ensurer.reset-size9.size2{font-size:0.34722222em;}.css-63uqft .katex .sizing.reset-size9.size3,.css-63uqft .katex .fontsize-ensurer.reset-size9.size3{font-size:0.40509259em;}.css-63uqft .katex .sizing.reset-size9.size4,.css-63uqft .katex .fontsize-ensurer.reset-size9.size4{font-size:0.46296296em;}.css-63uqft .katex .sizing.reset-size9.size5,.css-63uqft .katex .fontsize-ensurer.reset-size9.size5{font-size:0.52083333em;}.css-63uqft .katex .sizing.reset-size9.size6,.css-63uqft .katex .fontsize-ensurer.reset-size9.size6{font-size:0.5787037em;}.css-63uqft .katex .sizing.reset-size9.size7,.css-63uqft .katex .fontsize-ensurer.reset-size9.size7{font-size:0.69444444em;}.css-63uqft .katex .sizing.reset-size9.size8,.css-63uqft .katex .fontsize-ensurer.reset-size9.size8{font-size:0.83333333em;}.css-63uqft .katex .sizing.reset-size9.size9,.css-63uqft .katex .fontsize-ensurer.reset-size9.size9{font-size:1em;}.css-63uqft .katex .sizing.reset-size9.size10,.css-63uqft .katex .fontsize-ensurer.reset-size9.size10{font-size:1.20023148em;}.css-63uqft .katex .sizing.reset-size9.size11,.css-63uqft .katex .fontsize-ensurer.reset-size9.size11{font-size:1.43981481em;}.css-63uqft .katex .sizing.reset-size10.size1,.css-63uqft .katex .fontsize-ensurer.reset-size10.size1{font-size:0.24108004em;}.css-63uqft .katex .sizing.reset-size10.size2,.css-63uqft .katex .fontsize-ensurer.reset-size10.size2{font-size:0.28929605em;}.css-63uqft .katex .sizing.reset-size10.size3,.css-63uqft .katex .fontsize-ensurer.reset-size10.size3{font-size:0.33751205em;}.css-63uqft .katex .sizing.reset-size10.size4,.css-63uqft .katex .fontsize-ensurer.reset-size10.size4{font-size:0.38572806em;}.css-63uqft .katex .sizing.reset-size10.size5,.css-63uqft .katex .fontsize-ensurer.reset-size10.size5{font-size:0.43394407em;}.css-63uqft .katex .sizing.reset-size10.size6,.css-63uqft .katex .fontsize-ensurer.reset-size10.size6{font-size:0.48216008em;}.css-63uqft .katex .sizing.reset-size10.size7,.css-63uqft .katex .fontsize-ensurer.reset-size10.size7{font-size:0.57859209em;}.css-63uqft .katex .sizing.reset-size10.size8,.css-63uqft .katex .fontsize-ensurer.reset-size10.size8{font-size:0.69431051em;}.css-63uqft .katex .sizing.reset-size10.size9,.css-63uqft .katex .fontsize-ensurer.reset-size10.size9{font-size:0.83317261em;}.css-63uqft .katex .sizing.reset-size10.size10,.css-63uqft .katex .fontsize-ensurer.reset-size10.size10{font-size:1em;}.css-63uqft .katex .sizing.reset-size10.size11,.css-63uqft .katex .fontsize-ensurer.reset-size10.size11{font-size:1.19961427em;}.css-63uqft .katex .sizing.reset-size11.size1,.css-63uqft .katex .fontsize-ensurer.reset-size11.size1{font-size:0.20096463em;}.css-63uqft .katex .sizing.reset-size11.size2,.css-63uqft .katex .fontsize-ensurer.reset-size11.size2{font-size:0.24115756em;}.css-63uqft .katex .sizing.reset-size11.size3,.css-63uqft .katex .fontsize-ensurer.reset-size11.size3{font-size:0.28135048em;}.css-63uqft .katex .sizing.reset-size11.size4,.css-63uqft .katex .fontsize-ensurer.reset-size11.size4{font-size:0.32154341em;}.css-63uqft .katex .sizing.reset-size11.size5,.css-63uqft .katex .fontsize-ensurer.reset-size11.size5{font-size:0.36173633em;}.css-63uqft .katex .sizing.reset-size11.size6,.css-63uqft .katex .fontsize-ensurer.reset-size11.size6{font-size:0.40192926em;}.css-63uqft .katex .sizing.reset-size11.size7,.css-63uqft .katex .fontsize-ensurer.reset-size11.size7{font-size:0.48231511em;}.css-63uqft .katex .sizing.reset-size11.size8,.css-63uqft .katex .fontsize-ensurer.reset-size11.size8{font-size:0.57877814em;}.css-63uqft .katex .sizing.reset-size11.size9,.css-63uqft .katex .fontsize-ensurer.reset-size11.size9{font-size:0.69453376em;}.css-63uqft .katex .sizing.reset-size11.size10,.css-63uqft .katex .fontsize-ensurer.reset-size11.size10{font-size:0.83360129em;}.css-63uqft .katex .sizing.reset-size11.size11,.css-63uqft .katex .fontsize-ensurer.reset-size11.size11{font-size:1em;}.css-63uqft .katex .delimsizing.size1{font-family:KaTeX_Size1;}.css-63uqft .katex .delimsizing.size2{font-family:KaTeX_Size2;}.css-63uqft .katex .delimsizing.size3{font-family:KaTeX_Size3;}.css-63uqft .katex .delimsizing.size4{font-family:KaTeX_Size4;}.css-63uqft .katex .delimsizing.mult .delim-size1>span{font-family:KaTeX_Size1;}.css-63uqft .katex .delimsizing.mult .delim-size4>span{font-family:KaTeX_Size4;}.css-63uqft .katex .nulldelimiter{display:inline-block;width:0.12em;}.css-63uqft .katex .delimcenter{position:relative;}.css-63uqft .katex .op-symbol{position:relative;}.css-63uqft .katex .op-symbol.small-op{font-family:KaTeX_Size1;}.css-63uqft .katex .op-symbol.large-op{font-family:KaTeX_Size2;}.css-63uqft .katex .op-limits>.vlist-t{text-align:center;}.css-63uqft .katex .accent>.vlist-t{text-align:center;}.css-63uqft .katex .accent .accent-body{position:relative;}.css-63uqft .katex .accent .accent-body:not(.accent-full){width:0;}.css-63uqft .katex .overlay{display:block;}.css-63uqft .katex .mtable .vertical-separator{display:inline-block;min-width:1px;}.css-63uqft .katex .mtable .arraycolsep{display:inline-block;}.css-63uqft .katex .mtable .col-align-c>.vlist-t{text-align:center;}.css-63uqft .katex .mtable .col-align-l>.vlist-t{text-align:left;}.css-63uqft .katex .mtable .col-align-r>.vlist-t{text-align:right;}.css-63uqft .katex .svg-align{text-align:left;}.css-63uqft .katex svg{display:block;position:absolute;width:100%;height:inherit;fill:currentColor;stroke:currentColor;fill-rule:nonzero;fill-opacity:1;stroke-width:1;stroke-linecap:butt;stroke-linejoin:miter;stroke-miterlimit:4;stroke-dasharray:none;stroke-dashoffset:0;stroke-opacity:1;}.css-63uqft .katex svg path{stroke:none;}.css-63uqft .katex img{border-style:none;min-width:0;min-height:0;max-width:none;max-height:none;}.css-63uqft .katex .stretchy{width:100%;display:block;position:relative;overflow:hidden;}.css-63uqft .katex .stretchy::before,.css-63uqft .katex .stretchy::after{content:'';}.css-63uqft .katex .hide-tail{width:100%;position:relative;overflow:hidden;}.css-63uqft .katex .halfarrow-left{position:absolute;left:0;width:50.2%;overflow:hidden;}.css-63uqft .katex .halfarrow-right{position:absolute;right:0;width:50.2%;overflow:hidden;}.css-63uqft .katex .brace-left{position:absolute;left:0;width:25.1%;overflow:hidden;}.css-63uqft .katex .brace-center{position:absolute;left:25%;width:50%;overflow:hidden;}.css-63uqft .katex .brace-right{position:absolute;right:0;width:25.1%;overflow:hidden;}.css-63uqft .katex .x-arrow-pad{padding:0 0.5em;}.css-63uqft .katex .cd-arrow-pad{padding:0 0.55556em 0 0.27778em;}.css-63uqft .katex .x-arrow,.css-63uqft .katex .mover,.css-63uqft .katex .munder{text-align:center;}.css-63uqft .katex .boxpad{padding:0 0.3em 0 0.3em;}.css-63uqft .katex .fbox,.css-63uqft .katex .fcolorbox{box-sizing:border-box;border:0.04em solid;}.css-63uqft .katex .cancel-pad{padding:0 0.2em 0 0.2em;}.css-63uqft .katex .cancel-lap{margin-left:-0.2em;margin-right:-0.2em;}.css-63uqft .katex .sout{border-bottom-style:solid;border-bottom-width:0.08em;}.css-63uqft .katex .angl{box-sizing:border-box;border-top:0.049em solid;border-right:0.049em solid;margin-right:0.03889em;}.css-63uqft .katex .anglpad{padding:0 0.03889em 0 0.03889em;}.css-63uqft .katex .eqn-num::before{counter-increment:katexEqnNo;content:'(' counter(katexEqnNo) ')';}.css-63uqft .katex .mml-eqn-num::before{counter-increment:mmlEqnNo;content:'(' counter(mmlEqnNo) ')';}.css-63uqft .katex .mtr-glue{width:50%;}.css-63uqft .katex .cd-vert-arrow{display:inline-block;position:relative;}.css-63uqft .katex .cd-label-left{display:inline-block;position:absolute;right:calc(50% + 0.3em);text-align:left;}.css-63uqft .katex .cd-label-right{display:inline-block;position:absolute;left:calc(50% + 0.3em);text-align:right;}.css-63uqft .katex-display{display:block;margin:1em 0;text-align:center;}.css-63uqft .katex-display>.katex{display:block;white-space:nowrap;}.css-63uqft .katex-display>.katex>.katex-html{display:block;position:relative;}.css-63uqft .katex-display>.katex>.katex-html>.tag{position:absolute;right:0;}.css-63uqft .katex-display.leqno>.katex>.katex-html>.tag{left:0;right:auto;}.css-63uqft .katex-display.fleqn>.katex{text-align:left;padding-left:2em;}.css-63uqft body{counter-reset:katexEqnNo mmlEqnNo;}.css-63uqft table{width:-webkit-max-content;width:-moz-max-content;width:max-content;}.css-63uqft .tableBlock{max-width:100%;margin-bottom:1rem;overflow-y:scroll;}.css-63uqft .tableBlock thead,.css-63uqft .tableBlock thead th{border-bottom:1px solid #333!important;}.css-63uqft .tableBlock th,.css-63uqft .tableBlock td{padding:10px;text-align:left;}.css-63uqft .tableBlock th{font-weight:bold!important;}.css-63uqft .tableBlock caption{caption-side:bottom;color:#555;font-size:12px;font-style:italic;text-align:center;}.css-63uqft .tableBlock caption>p{margin:0;}.css-63uqft .tableBlock th>p,.css-63uqft .tableBlock td>p{margin:0;}.css-63uqft .tableBlock [data-background-color='aliceblue']{background-color:#f0f8ff;color:#000;}.css-63uqft .tableBlock [data-background-color='black']{background-color:#000;color:#fff;}.css-63uqft .tableBlock [data-background-color='chocolate']{background-color:#d2691e;color:#fff;}.css-63uqft .tableBlock [data-background-color='cornflowerblue']{background-color:#6495ed;color:#fff;}.css-63uqft .tableBlock [data-background-color='crimson']{background-color:#dc143c;color:#fff;}.css-63uqft .tableBlock [data-background-color='darkblue']{background-color:#00008b;color:#fff;}.css-63uqft .tableBlock [data-background-color='darkseagreen']{background-color:#8fbc8f;color:#000;}.css-63uqft .tableBlock [data-background-color='deepskyblue']{background-color:#00bfff;color:#000;}.css-63uqft .tableBlock [data-background-color='gainsboro']{background-color:#dcdcdc;color:#000;}.css-63uqft .tableBlock [data-background-color='grey']{background-color:#808080;color:#fff;}.css-63uqft .tableBlock [data-background-color='lemonchiffon']{background-color:#fffacd;color:#000;}.css-63uqft .tableBlock [data-background-color='lightpink']{background-color:#ffb6c1;color:#000;}.css-63uqft .tableBlock [data-background-color='lightsalmon']{background-color:#ffa07a;color:#000;}.css-63uqft .tableBlock [data-background-color='lightskyblue']{background-color:#87cefa;color:#000;}.css-63uqft .tableBlock [data-background-color='mediumblue']{background-color:#0000cd;color:#fff;}.css-63uqft .tableBlock [data-background-color='omnigrey']{background-color:#f0f0f0;color:#000;}.css-63uqft .tableBlock [data-background-color='white']{background-color:#fff;color:#000;}.css-63uqft .tableBlock [data-text-align='center']{text-align:center;}.css-63uqft .tableBlock [data-text-align='left']{text-align:left;}.css-63uqft .tableBlock [data-text-align='right']{text-align:right;}.css-63uqft .tableBlock [data-vertical-align='bottom']{vertical-align:bottom;}.css-63uqft .tableBlock [data-vertical-align='middle']{vertical-align:middle;}.css-63uqft .tableBlock [data-vertical-align='top']{vertical-align:top;}.css-63uqft .tableBlock__font-size--xxsmall{font-size:10px;}.css-63uqft .tableBlock__font-size--xsmall{font-size:12px;}.css-63uqft .tableBlock__font-size--small{font-size:14px;}.css-63uqft .tableBlock__font-size--large{font-size:18px;}.css-63uqft .tableBlock__border--some tbody tr:not(:last-child){border-bottom:1px solid #e2e5e7;}.css-63uqft .tableBlock__border--bordered td,.css-63uqft .tableBlock__border--bordered th{border:1px solid #e2e5e7;}.css-63uqft .tableBlock__border--borderless tbody+tbody,.css-63uqft .tableBlock__border--borderless td,.css-63uqft .tableBlock__border--borderless th,.css-63uqft .tableBlock__border--borderless tr,.css-63uqft .tableBlock__border--borderless thead,.css-63uqft .tableBlock__border--borderless thead th{border:0!important;}.css-63uqft .tableBlock:not(.tableBlock__table-striped) tbody tr{background-color:unset!important;}.css-63uqft .tableBlock__table-striped tbody tr:nth-of-type(odd){background-color:#f9fafc!important;}.css-63uqft .tableBlock__table-compactl th,.css-63uqft .tableBlock__table-compact td{padding:3px!important;}.css-63uqft .tableBlock__full-size{width:100%;}.css-63uqft .textBlock{margin-bottom:16px;}.css-63uqft .textBlock__text-formatting--finePrint{font-size:12px;}.css-63uqft .textBlock__text-infoBox{padding:0.75rem 1.25rem;margin-bottom:1rem;border:1px solid transparent;border-radius:0.25rem;}.css-63uqft .textBlock__text-infoBox p{margin:0;}.css-63uqft .textBlock__text-infoBox--primary{background-color:#cce5ff;border-color:#b8daff;color:#004085;}.css-63uqft .textBlock__text-infoBox--secondary{background-color:#e2e3e5;border-color:#d6d8db;color:#383d41;}.css-63uqft .textBlock__text-infoBox--success{background-color:#d4edda;border-color:#c3e6cb;color:#155724;}.css-63uqft .textBlock__text-infoBox--danger{background-color:#f8d7da;border-color:#f5c6cb;color:#721c24;}.css-63uqft .textBlock__text-infoBox--warning{background-color:#fff3cd;border-color:#ffeeba;color:#856404;}.css-63uqft .textBlock__text-infoBox--info{background-color:#d1ecf1;border-color:#bee5eb;color:#0c5460;}.css-63uqft .textBlock__text-infoBox--dark{background-color:#d6d8d9;border-color:#c6c8ca;color:#1b1e21;}.css-63uqft .text-overline{-webkit-text-decoration:overline;text-decoration:overline;}.css-63uqft.css-63uqft{color:#2B3148;background-color:transparent;font-family:var(--calculator-ui-font-family),Verdana,sans-serif;font-size:20px;line-height:24px;overflow:visible;padding-top:0px;position:relative;}.css-63uqft.css-63uqft:after{content:'';-webkit-transform:scale(0);-moz-transform:scale(0);-ms-transform:scale(0);transform:scale(0);position:absolute;border:2px solid #EA9430;border-radius:2px;inset:-8px;z-index:1;}.css-63uqft .js-external-link-button.link-like,.css-63uqft .js-external-link-anchor{color:inherit;border-radius:1px;-webkit-text-decoration:underline;text-decoration:underline;}.css-63uqft .js-external-link-button.link-like:hover,.css-63uqft .js-external-link-anchor:hover,.css-63uqft .js-external-link-button.link-like:active,.css-63uqft .js-external-link-anchor:active{text-decoration-thickness:2px;text-shadow:1px 0 0;}.css-63uqft .js-external-link-button.link-like:focus-visible,.css-63uqft .js-external-link-anchor:focus-visible{outline:transparent 2px dotted;box-shadow:0 0 0 2px #6314E6;}.css-63uqft p,.css-63uqft div{margin:0;display:block;}.css-63uqft pre{margin:0;display:block;}.css-63uqft pre code{display:block;width:-webkit-fit-content;width:-moz-fit-content;width:fit-content;}.css-63uqft pre:not(:first-child){padding-top:8px;}.css-63uqft ul,.css-63uqft ol{display:block margin:0;padding-left:20px;}.css-63uqft ul li,.css-63uqft ol li{padding-top:8px;}.css-63uqft ul ul,.css-63uqft ol ul,.css-63uqft ul ol,.css-63uqft ol ol{padding-top:0;}.css-63uqft ul:not(:first-child),.css-63uqft ol:not(:first-child){padding-top:4px;} Test setup
Choose test type
t-test for the population mean, μ, based on one independent sample . Null hypothesis H 0 : μ = μ 0
Alternative hypothesis H 1
Test details
Significance level α
The probability that we reject a true H 0 (type I error).
Degrees of freedom
Calculated as sample size minus one.
Test results
Hypothesis Testing Calculator Online
Hypothesis testing is a foundational method used in statistics to infer the validity of a hypothesis about a population parameter. The Hypothesis Testing Calculator facilitates this process by automating the computations necessary for the t-test , a method used to compare sample means against a hypothesized mean or against each other. Let’s delve into the formulas this calculator uses to execute one-sample and two-sample t-tests.
One-Sample t-Test
This test is used to determine if the mean (x̄) of your sample is statistically different from a hypothesized population mean (μ₀).
Two-Sample t-Test
Equal variances:, unequal variances (welch’s t-test):.
t = (x̄₁ – x̄₂) / (√((s₁² / n₁) + (s₂² / n₂)))
Table of Critical t-Values
| Confidence Level (%) | df=10 | df=30 | df=50 | df=100 |
|---|---|---|---|---|
| 90 | 1.812 | 1.697 | 1.676 | 1.660 |
| 95 | 2.228 | 2.042 | 2.009 | 1.984 |
| 99 | 3.169 | 2.750 | 2.678 | 2.626 |
These values are crucial in hypothesis testing as they help define the threshold for significance, assisting users of the calculator in interpreting their results accurately.
Using the one-sample t-test formula:
t = (74 – 70) / (8 / √36) = (4 / 1.333) = 3.00
Most Common FAQs
The p-value represents the probability of obtaining test results at least as extreme as the results observed, under the assumption that the null hypothesis is correct. A low p-value (typically below 0.05) indicates strong evidence against the null hypothesis, hence it is usually rejected.
Yes, while the t-test is specifically design for means, the principles of hypothesis testing apply to other parameters such as proportions and variances. Which can also be tested using appropriate versions of hypothesis tests such as the z-test and F-test.
Related Calculators
Leave a comment cancel reply.
For optimal use, please visit DATAtab on your desktop PC!
Metric Variables:
Ordinal variables:, nominal variables:, hypothesis test calculator.
Do you want to calculate a hypothesis test such as a t-test , Chi Square test or an ANOVA ? You can do that easily here in the browser.
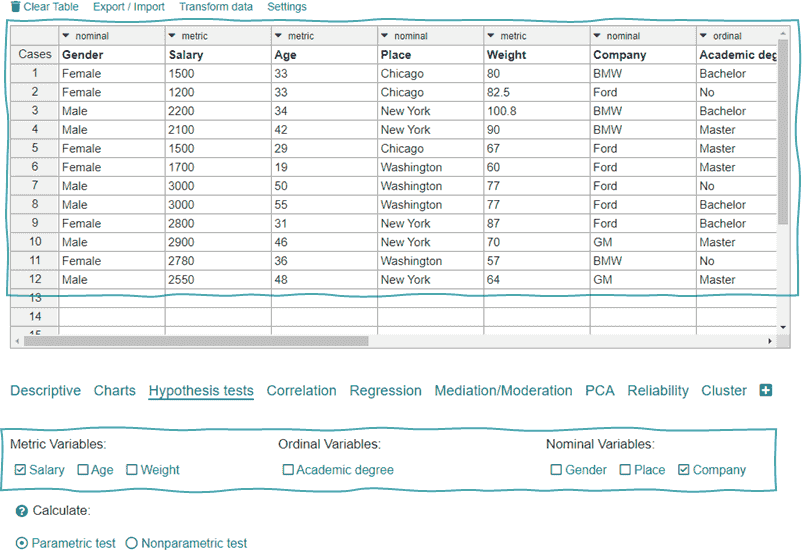
If you want to use your own data just clear the upper table
- Clear the table in the Hypothesis test calculator.
- Copy your data into the table.
- Select the variables.
In the hypothesis test calculator you can calculate e.g. a t-test, a chi-square test, a binomial test or an analysis of variance. If you need a more detailed explanation, you can find more information in the tutorials.
In order to use the hypothesis test calculator, you must first formulate your hypothesis and collect your data. DATAtab will then suggest the hypothesis test you need based on the data entered into the statistics calculator.
p value calculator
With the p value calculator you can calculate the p value for different tests. There is a wide range of methods for this. Just click on the variables you want to evaluate above and DATAtab will give you the tests you can use.
For example, if you select a metric and a categorical variable, the Independent t-Test calculator is automatically selected. If your data is not normally distributed, simply use the Mann-Whitney U-test calculator.
H0 and H1 calculator
With the h0 and h1 calculator for the different hypothesis test you can calculate the p-value which gives you an indication if you can reject the H0 or not.
Cite DATAtab: DATAtab Team (2024). DATAtab: Online Statistics Calculator. DATAtab e.U. Graz, Austria. URL https://datatab.net

- Calculators
Copyright © 2024 All Rights Reserved. Made by 🐒cibey with ❤️
Hypothesis Testing Calculator
Enhanced hypothesis test calculator, hypothesis test calculator: your essential tool for statistical analysis.
The Hypothesis Test Calculator is an intuitive, web-based tool crafted to simplify the process of conducting statistical hypothesis tests. Whether you’re a researcher, student, or educator, this tool streamlines the process, providing accurate results and clear visualizations with minimal effort.
How to Use the Hypothesis Test Calculator
- Step 1: Start by entering the required data into the respective input fields. You’ll need to provide the sample mean, sample size, population mean, population standard deviation, and the significance level (alpha). If you have raw data, you can also upload a CSV file directly.
- Step 2: Choose whether you’re conducting a one-tailed or two-tailed test. If you’re interested in knowing the confidence interval, select the option to calculate it.
- Step 3: Click the “Perform Test” button. The tool will quickly process the information, performing the hypothesis test and generating a detailed report.
- Step 4: Review the results. The calculator provides the calculated t-value, critical value, and the test’s conclusion. You can also visualize the sample distribution with a dynamic chart that compares it to the population mean.
Understanding Hypothesis Testing
Hypothesis testing is a core statistical method used to determine whether there is enough evidence in a sample to infer that a certain condition is true for the entire population. The test compares the sample data against the null hypothesis, which typically represents the status quo or a baseline condition.
In this context, the Hypothesis Test Calculator is particularly useful for t-tests, which assess whether the sample mean significantly differs from a known population mean. By inputting your data, the tool computes the necessary values, helping you determine whether to reject the null hypothesis.
Hypothesis Test Formula:
$$ t = \frac{\text{Sample Mean} – \text{Population Mean}}{\text{Standard Error}} $$
Features and Advantages
- User-Friendly Design: The calculator is built with simplicity in mind, allowing users to quickly enter data and obtain results without any hassle.
- Rapid Calculations: With just a few clicks, the tool performs complex statistical calculations, delivering results almost instantaneously.
- Comprehensive Results: Not only does the calculator provide the final conclusion, but it also breaks down the steps involved in the calculation, aiding in understanding the statistical process.
- Visual Representation: The tool includes a dynamic chart that illustrates the sample distribution against the population mean, helping users visualize the data and results.
- Versatile Input Options: You can input data manually or upload a CSV file, making it easy to work with real datasets.
- Educational Utility: Perfect for both students and educators, the Hypothesis Test Calculator serves as an excellent learning tool, offering insights into the mechanics of hypothesis testing.
Enhancing Your Statistical Toolkit
The Hypothesis Test Calculator is an invaluable resource for anyone involved in data analysis. It simplifies the hypothesis testing process, delivering clear, accurate results and visual aids that enhance your understanding of statistical concepts.
Try the Hypothesis Test Calculator today and take your statistical analysis to the next level!
How useful was this Tool?
Click on a star to rate it!
Average rating / 5. Vote count:
No votes so far! Be the first to rate this post.
As you found this tool useful...
Share it on social media!
We are sorry that this tool was not useful for you!
Let us improve this tool!
Tell us how we can improve this tool?
Related Tools:
- Website Responsive Preview Testing Tool
- ANOVA Calculator
- Time Duration Calculator
- Protein Calculator
- Biweekly Payment Calculator
- Meal planning and recipe calculator
Sign in to your account
Username or Email Address
Remember Me

Hypothesis Testing Calculator
Understanding Hypothesis Testing: A Guide to the Hypothesis Testing Calculator
Hypothesis testing is a crucial statistical method used to make informed decisions about data and draw conclusions. Whether you’re a student, researcher, or professional, a Hypothesis Testing Calculator can be an invaluable tool in your statistical toolkit. Let’s explore what hypothesis testing is and how this calculator can assist you:
Hypothesis Testing Basics:
- Null Hypothesis (H0): This is the default assumption or claim that there is no significant difference or effect. It’s often denoted as H0.
- Alternative Hypothesis (Ha): This is the statement that contradicts the null hypothesis. It suggests that there is a significant difference or effect. It’s denoted as Ha.
- Significance Level (α): This is the predetermined threshold (e.g., 0.05 or 5%) used to determine statistical significance. If the calculated p-value is less than α, you reject the null hypothesis.
- p-value: This is the probability of observing the results (or more extreme results) if the null hypothesis is true. A small p-value suggests that the results are unlikely under the null hypothesis.
Key Features of the Hypothesis Testing Calculator:
- Input Parameters: The calculator typically requires you to input sample data, choose the type of test (e.g., t-test, chi-square test), specify the null and alternative hypotheses, and set the significance level.
- Calculations: Once you input the data and parameters, the calculator performs the necessary statistical tests and calculations. It generates results such as the test statistic, degrees of freedom, and the p-value.
- Interpretation: Based on the results, the calculator helps you determine whether to reject or fail to reject the null hypothesis. It provides an interpretation of the findings, which is crucial for drawing conclusions.
- Visual Representation: Some calculators may offer visual aids like graphs or charts to help you better understand the data distribution and test results.
Significance of the Hypothesis Testing Calculator:
- Scientific Research: Researchers across various fields use hypothesis testing to validate their hypotheses and draw meaningful conclusions from data.
- Quality Control: Industries use hypothesis testing to ensure the quality and consistency of products and processes.
- Medical Studies: In medical research, hypothesis testing helps assess the effectiveness of treatments or interventions.
- Academics: Students and educators use hypothesis testing to teach and learn statistical concepts and conduct experiments.
- Data-Driven Decisions: Businesses use hypothesis testing to make data-driven decisions, such as whether to launch a new product based on market research.
Conclusion:
The Hypothesis Testing Calculator is a powerful tool that simplifies complex statistical analysis and enables data-driven decision-making. Whether you’re conducting experiments, analyzing survey data, or performing quality control, understanding hypothesis testing and using this calculator can help you make informed choices and contribute to evidence-based research and decision-making.
One Sample T Test Calculator
Enter sample data
Reporting results in APA style
One sample t-test, what is a one sample t-test, how to use the one sample t test calculator, calculators.
Two sample t test
Kruskal wallis
Pearson product moment
Two sample f test
One way anova
Bartlett test
Chi square goodness of fit
Fligner killeen
Hypothesis Test Calculator
Upload your data set below to get started
Or input your data as csv
Sharing helps us build more free tools

- Calculators
- Descriptive Statistics
- Merchandise
- Which Statistics Test?
Single Sample T-Test Calculator
A single sample t-test (or one sample t-test) is used to compare the mean of a single sample of scores to a known or hypothetical population mean. So, for example, it could be used to determine whether the mean diastolic blood pressure of a particular group differs from 85, a value determined by a previous study.
Requirements
- The data is normally distributed
- Scale of measurement should be interval or ratio
- A randomized sample from a defined population
Null Hypothesis
H 0 : M - μ = 0, where M is the sample mean and μ is the population or hypothesized mean.
As above, the null hypothesis is that there is no difference between the sample mean and the known or hypothesized population mean.

t-Test Calculator
T-test - work with steps.
Input Data : Data set x = 3, 11, 17, 28, 34 Data set y = 5, 8, 13, 19, 28 Total number of elements = 5 Objective : Find the t-score by using mean and standard deviation. Solution : Mean 1 = (3 + 11 + 17 + 28 + 34)/5 = 93/5 Mean 1 = 18.6 Mean 2 = (5 + 8 + 13 + 19 + 28)/5 = 73/5 Mean 2 = 14.6 SD1 = √(1/5 - 1) x ((3 - 18.6) 2 + ( 11 - 18.6) 2 + ( 17 - 18.6) 2 + ( 28 - 18.6) 2 + ( 34 - 18.6) 2 ) = √(1/4) x ((-15.6) 2 + (-7.6) 2 + (-1.6) 2 + (9.4) 2 + (15.4) 2 ) = √(0.25) x ((243.36) + (57.76) + (2.56) + (88.36) + (237.16)) = √(0.25) x 629.2 = √157.3 SD1 = 12.5419 SD2 = √(1/5 - 1) x ((5 - 14.6) 2 + ( 8 - 14.6) 2 + ( 13 - 14.6) 2 + ( 19 - 14.6) 2 + ( 28 - 14.6) 2 ) = √(1/4) x ((-9.6) 2 + (-6.6) 2 + (-1.6) 2 + (4.4) 2 + (13.4) 2 ) = √(0.25) x ((92.16) + (43.56) + (2.56) + (19.36) + (179.56)) = √(0.25) x 337.2 = √84.3 SD2 = 9.1815 t-score = x 1 - x 2 √(SD1 2 /n1 + SD2 2 /n2) = 18.6 - 14.6 √((12.5419) 2 /5 + (9.1815) 2 /5) = 4 √((157.3)/5 + (84.3)/5) = 4 √(31.46 + 16.86) = 4 √(48.32) = 4 6.9513 t-score = 0.5754
What is t-Test?
A hypothesis test consists of two hypotheses, the null hypothesis and the alternative hypothesis or research hypothesis. The symbol $H_0$ represents the null hypothesis. The null hypothesis reflects that there will be no observed effect on the experiment. The null hypothesis consists of an equal sign. The alternative hypothesis reflects that there is an observed effect on the experiment. The symbol $H_a$ represents the alternative hypothesis. The first step in testing is to determine the null hypothesis and the alternative hypothesis. Regarding the testing hypothesis, there are some important terms. Rejection region is the set of values leads to rejection of the null hypothesis. Non-rejection region is the set of values that leads to nonrejection of the null hypothesis. Critical values are the value that separates the rejection and non-rejection regions. The t-Test is used in comparing the means of two populations. There are two approaches:
How to Find t-Critical Value
T-test with mean and standard deviation.
A t-Test is one of the most frequently used tests in statistics. A t-Test is useful to conclude if the results are correct and applicable to the entire population. If we want to analyze simple experiments or when making simple comparisons between levels of independent variable we use the t-Test. It's used in comparison between two separate groups of individuals, for example: male vs female, experimental vs control group, etc. Practice Problem 1: There are two company A and B. We want to test average age of employees at these companies so we use a random sample of employee ages from each company.
| Company A | Company B | |
| Mean | 43.2 | 36.7 |
| Standard Deviation | 7 | 8.3 |
| Number of Employess | 50 | 66 |
| Serbia | United States | |
| Mean | 43.2 | 5.2 |
| Standard Deviation | 1.2 | 8.3 |
| Number of Employess | 67 | 166 |
- Math Lessons
- Math Formulas
- Calculators
Math Calculators, Lessons and Formulas
It is time to solve your math problem
- HW Help (paid service)
- Statistics and probability
- T-test calculator
T-Test calculator
The Student's t-test is used to determine if means of two data sets differ significantly. This calculator will generate a step by step explanation on how to apply t – test.
|























IMAGES
VIDEO
COMMENTS
Hypothesis Testing Calculator. The first step in hypothesis testing is to calculate the test statistic. The formula for the test statistic depends on whether the population standard deviation (σ) is known or unknown. If σ is known, our hypothesis test is known as a z test and we use the z distribution. If σ is unknown, our hypothesis test is ...
Calculation Example: There are six steps you would follow in hypothesis testing: Formulate the null and alternative hypotheses in three different ways: H 0: θ = θ 0 v e r s u s H 1: θ ≠ θ 0. H 0: θ ≤ θ 0 v e r s u s H 1: θ> θ 0. H 0: θ ≥ θ 0 v e r s u s H 1: θ <θ 0.
How to Use Our Hypothesis Testing Calculator. Using our hypothesis testing calculator is straightforward. Simply select the type of test you need from the list above, input your data, and follow the prompts. Our calculators will guide you through each step, ensuring you understand the process and obtain accurate results.
t-test Calculator | Formula | p-value
Select Data Type: Choose whether to input summary statistics directly or provide a data set. Input Your Data: Enter the required values such as population mean, sample size, sample mean, and sample standard deviation. Set Hypotheses: Specify the null and alternative hypotheses. Calculate: Click the "Calculate" button to see the test statistic ...
Online Statistics Calculator: Hypothesis testing, t-test, chi ...
Hypothesis testing is a foundational method used in statistics to infer the validity of a hypothesis about a population parameter. The Hypothesis Testing Calculator facilitates this process by automating the computations necessary for the t-test, a method used to compare sample means against a hypothesized mean or against each other.Let's delve into the formulas this calculator uses to ...
Copy your data into the table. Select the variables. In the hypothesis test calculator you can calculate e.g. a t-test, a chi-square test, a binomial test or an analysis of variance. If you need a more detailed explanation, you can find more information in the tutorials. In order to use the hypothesis test calculator, you must first formulate ...
The Hypothesis Test Calculator is an invaluable resource for anyone involved in data analysis. It simplifies the hypothesis testing process, delivering clear, accurate results and visual aids that enhance your understanding of statistical concepts. Try the Hypothesis Test Calculator today and take your statistical analysis to the next level!
Calculations: Once you input the data and parameters, the calculator performs the necessary statistical tests and calculations. It generates results such as the test statistic, degrees of freedom, and the p-value. Interpretation: Based on the results, the calculator helps you determine whether to reject or fail to reject the null hypothesis.
Hypothesis Test for a Population Mean With Data Calculator. Type in the values from the data set separated by commas, for example, 2,4,5,8,11,2. Then type in the population standard deviation σ σ if it is known or leave it blank if it is not known. Then choose a left, right or two tailed test, and the hypothesized mean.
One Sample T Test Calculator
Hypothesis Test Calculator. Upload your data set below to get started. Upload File. Or input your data as csv. column_one,column_two,column_three 1,2,3 4,5,6 7,8,9. Submit CSV. Sharing helps us build more free tools.
T-Test Calculator for 2 Independent Means
Single Sample T-Test Calculator. A single sample t-test (or one sample t-test) is used to compare the mean of a single sample of scores to a known or hypothetical population mean. So, for example, it could be used to determine whether the mean diastolic blood pressure of a particular group differs from 85, a value determined by a previous study.
t-test calculator is an online statistics tool to estimate the significance of observed differences between the means of two samples when there is a null hypothesis that is no significant difference between the means by using standard deviation. It is necessary to follow the next steps: Enter two samples (observed values) in the box. These values must be real numbers or variables and may be ...
Two sample and one sample t-test calculator with step by step explanation. Site map; Math Tests; Math Lessons; Math Formulas; Calculators; Math Calculators, Lessons and Formulas ... Twelve younger adults and twelve older adults conducted a life satisfaction test. The data are presented in the table below. Compute the appropriate t-test ...
A one sample t-test is used to test whether or not the mean of a population is equal to some value. To perform a one sample t-test, simply fill in the information below and then click the "Calculate" button. Enter raw data Enter summary data. 301, 298, 295, 297, 304, 305, 309, 298, 291, 299, 293, 304. μ0 (hypothesized population mean) t ...
Test Statistic Calculator. Choose the method, enter the values into the test statistic calculator, and click on the "Calculate" button to calculate the statistical value for hypothesis evaluation. Select Format. Enter the data in row. Enter mean, SEM and n. Enter mean, SD and n. Group One.
Two Sample t-test Calculator
These calculators are valuable tools for students, researchers, and professionals needing to make data-driven decisions. Our site provides various statistics calculators to help you with your statistical analysis needs, including critical value, p-value, hypothesis testing, regression analysis, and descriptive statistics calculators.
If the two population variances are assumed to be equal, an alternative formula for computing the degrees of freedom is used. It's simply df = n1 + n2 - 2. This is a simple extension of the formula for the one population case. In the one population case the degrees of freedom is given by df = n - 1. If we add up the degrees of freedom for the ...
Hypothesis testing is a way to use data to check if your guesses or assumptions are likely true. By following these steps—setting up your hypotheses, choosing the right test, deciding on a significance level, analyzing your data, finding the p-value, making a decision, and reporting results—you can determine if your data supports or ...
Two Sample T-Test Calculator
Introduction. Understanding the relationship between sampling distributions, probability distributions, and hypothesis testing is the crucial concept in the NHST — Null Hypothesis Significance Testing — approach to inferential statistics. is crucial, and many introductory text books are excellent here. I will add some here to their discussion, perhaps with a different approach, but the ...
Hypothesis testing is a statistical method used to draw conclusions about populations from sample data, typically represented in tables. With the prevalence of graph representations in real-life applications, hypothesis testing on graphs is gaining importance. In this work, we formalize node, edge, and path hypotheses on attributed graphs.
A more complex model with metabolic pathways is needed to test most predictions of the screening hypothesis. However, we can test one prediction: Based on the screening hypothesis, plants with more metabolites should have a higher probability of having a metabolite that is active against a newly colonizing herbivore and therefore a higher ...
Background Cardiovascular toxicity represents a significant adverse consequence of cancer therapies, yet there remains a paucity of effective biomarkers for its timely monitoring and diagnosis. To give a first evidence able to elucidate the role of Growth Differentiation Factor 15 (GDF15) in the context of cancer diagnosis and its specific association with cardiac indicators in cancer patients ...