- Biology Article
- Recombinant Dna Technology

Recombinant DNA Technology
A technique mainly used to change the phenotype of an organism (host) when a genetically altered vector is introduced and integrated into the genome of the organism. So, basically, this process involves the introduction of a foreign piece of DNA structure into the genome which contains our gene of interest. This gene which is introduced is the recombinant gene and the technique is called the recombinant DNA technology.
There are multiple steps, tools and other specific procedures followed in the recombinant DNA technology, which is used for producing artificial DNA to generate the desired product. Let’s understand each step more in detail.
Table of Contents
- Explanation
- Application
DNA Cloning
Applications of gene cloning, what is recombinant dna technology.
The technology used for producing artificial DNA through the combination of different genetic materials (DNA) from different sources is referred to as Recombinant DNA Technology. Recombinant DNA technology is popularly known as genetic engineering.
The recombinant DNA technology emerged with the discovery of restriction enzymes in the year 1968 by Swiss microbiologist Werner Arber,
Inserting the desired gene into the genome of the host is not as easy as it sounds. It involves the selection of the desired gene for administration into the host followed by a selection of the perfect vector with which the gene has to be integrated and recombinant DNA formed.
Thus the recombinant DNA has to be introduced into the host. And at last, it has to be maintained in the host and carried forward to the offspring.
Also Refer- Genes
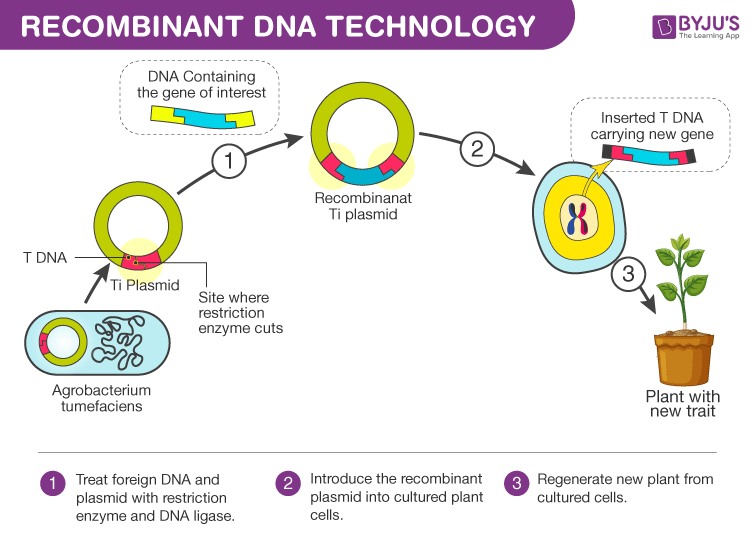
Tools Of Recombinant DNA Technology
The enzymes which include the restriction enzymes help to cut, the polymerases- help to synthesize and the ligases- help to bind. The restriction enzymes used in recombinant DNA technology play a major role in determining the location at which the desired gene is inserted into the vector genome. They are two types, namely Endonucleases and Exonucleases.
The Endonucleases cut within the DNA strand whereas the Exonucleases remove the nucleotides from the ends of the strands. The restriction endonucleases are sequence-specific which are usually palindrome sequences and cut the DNA at specific points. They scrutinize the length of DNA and make the cut at the specific site called the restriction site. This gives rise to sticky ends in the sequence. The desired genes and the vectors are cut by the same restriction enzymes to obtain the complementary sticky notes, thus making the work of the ligases easy to bind the desired gene to the vector.
The vectors – help in carrying and integrating the desired gene. These form a very important part of the tools of recombinant DNA technology as they are the ultimate vehicles that carry forward the desired gene into the host organism. Plasmids and bacteriophages are the most common vectors in recombinant DNA technology that are used as they have a very high copy number. The vectors are made up of an origin of replication- This is a sequence of nucleotides from where the replication starts, a selectable marker – constitute genes which show resistance to certain antibiotics like ampicillin; and cloning sites – the sites recognized by the restriction enzymes where desired DNAs are inserted.
Host organism – into which the recombinant DNA is introduced. The host is the ultimate tool of recombinant DNA technology which takes in the vector engineered with the desired DNA with the help of the enzymes.
There are a number of ways in which these recombinant DNAs are inserted into the host, namely – microinjection, biolistics or gene gun, alternate cooling and heating, use of calcium ions, etc.
Also Read: Bioinformatics
Process of Recombinant DNA Technology
The complete process of recombinant DNA technology includes multiple steps, maintained in a specific sequence to generate the desired product.
Step-1. Isolation of Genetic Material.
The first and the initial step in Recombinant DNA technology is to isolate the desired DNA in its pure form i.e. free from other macromolecules.
Step-2. Cutting the gene at the recognition sites.
The restriction enzymes play a major role in determining the location at which the desired gene is inserted into the vector genome. These reactions are called ‘restriction enzyme digestions’.
Step-3. Amplifying the gene copies through Polymerase chain reaction (PCR).
It is a process to amplify a single copy of DNA into thousands to millions of copies once the proper gene of interest has been cut using restriction enzymes.
Step-4. Ligation of DNA Molecules.
In this step of Ligation, the joining of the two pieces – a cut fragment of DNA and the vector together with the help of the enzyme DNA ligase.
Step-5. Insertion of Recombinant DNA Into Host.
In this step, the recombinant DNA is introduced into a recipient host cell. This process is termed as Transformation. Once the recombinant DNA is inserted into the host cell, it gets multiplied and is expressed in the form of the manufactured protein under optimal conditions.
As mentioned in Tools of recombinant DNA technology, there are various ways in which this can be achieved. The effectively transformed cells/organisms carry forward the recombinant gene to the offspring.
Also Read: R-Factor
Application of Recombinant DNA Technology
- DNA technology is also used to detect the presence of HIV in a person.
- Gene Therapy – It is used as an attempt to correct the gene defects which give rise to heredity diseases.
- Clinical diagnosis – ELISA is an example where the application of recombinant
- Recombinant DNA technology is widely used in Agriculture to produce genetically-modified organisms such as Flavr Savr tomatoes, golden rice rich in proteins, and Bt-cotton to protect the plant against ball worms and a lot more.
- In the field of medicines, Recombinant DNA technology is used for the production of Insulin.
Also Refer: Genetically Modified Organisms (GMO)
A clone is a cluster of individual entities or cells that are descended from one progenitor. Clones are genetically identical as the cell simply replicates producing identical daughter cells every time. Scientists are able to generate multiple copies of a single fragment of DNA, a gene which can be used to create identical copies constituting a DNA clone. DNA cloning takes place through the insertion of DNA fragments into a tiny DNA molecule. This molecule is made to replicate within a living cell, for instance, a bacterium. The tiny replicating molecule is known as the carrier of the DNA vector.
Yeast cells, viruses, and Plasmids are the most commonly used vectors. Plasmids are circular DNA molecules that are introduced from bacteria. They are not part of the main cellular genome. It carries genes, which provide the host cell with beneficial properties such as mating ability, and drug resistance. They can be conveniently manipulated as they are small enough and they are capable of carrying extra DNA which is weaved into them.
Explore more: Genetic Disorders.
Listed below are the applications of gene cloning:
- Gene Cloning plays an important role in the medicinal field. It is used in the production of hormones, vitamins and antibiotics.
- Gene cloning finds its applications in the agricultural field. Nitrogen fixation is carried out by cyanobacteria wherein desired genes can be used to enhance the productivity of crops and improvement of health. This practice reduces the use of fertilizers hence chemical-free produce is generated
- It can be applied to the science of identifying and detecting a clone containing a particular gene which can be manipulated by growing in a controlled environment
- It is used in gene therapy where a faulty gene is replaced by the insertion of a healthy gene. Medical ailments such as leukaemia and sickle cell anaemia can be treated with this principle.
Also Refer- Gene Therapy.
Frequently Asked Questions
Explain the roles of the following: (a) restriction enzymes (b) plasmids, explain pcr., discuss the applications of recombination from the point of view of genetic engineering..
- For the production of vaccines like the hepatitis B vaccine.
- Production of transgenic plants with improved qualities like insect and drought resistance and nutritional enrichment.
- Therapeutic protein production like insulin.
- Gene therapy in diseases like cancer, SCID etc.
- Production of transgenic animals with improved quality of milk and egg.

Put your understanding of this concept to test by answering a few MCQs. Click ‘Start Quiz’ to begin!
Select the correct answer and click on the “Finish” button Check your score and answers at the end of the quiz
Visit BYJU’S for all Biology related queries and study materials
Your result is as below
Request OTP on Voice Call
Leave a Comment Cancel reply
Your Mobile number and Email id will not be published. Required fields are marked *
Post My Comment
Thanks it is useful to send as a lecture notes during lock down period
Thank you so much. This module is really of great help in preparing an online class with my students.
- Share Share
Register with BYJU'S & Download Free PDFs
Register with byju's & watch live videos.


- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

1.11: Recombinant DNA Technology
- Last updated
- Save as PDF
- Page ID 73676

- Marjorie Hanneman, Walter Suza, Donald Lee, & Patricia Hain
- Iowa State University via Iowa State University Digital Press
Learning Objectives
- Understand the importance of recombinant DNA technology.
- Learn isolation of DNA and its separation on an agarose gel.
- Understand restriction and ligase enzymes and their application in gene cloning.
- Understand vectors and their application in gene cloning and expression.
- Understand polymerase chain termination reaction (PCR).
Introduction
Recombinant DNA (rDNA) technology has resulted in breakthroughs in crop and animal biotechnology. The power of rDNA technology comes from our ability to study and modify gene function by manipulating genes and transform them into cells of plant and animals. To arrive at this several tools of molecular biology are used including, DNA isolation and analysis, molecular cloning, quantification of gene expression, determination of gene copy number, transformation of the appropriate host for replication or transfer into crop plants and analyses of transgenic plants.
Definition and background
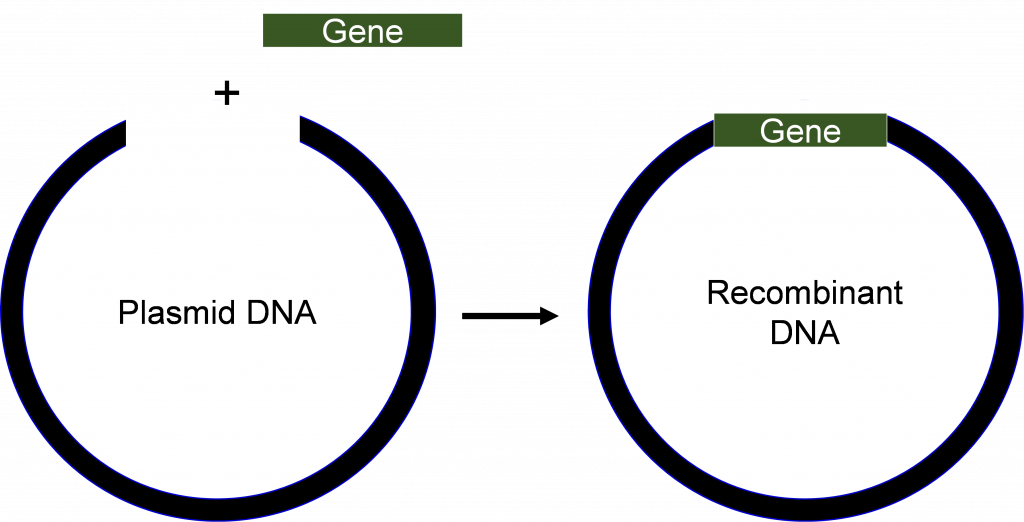
Recombinant rDNA technology involves procedures for analyzing or combining DNA fragments from one or several organisms (Figure 1) including the introduction of the rDNA molecule into a cell for its replication, or integration into the genome of the target cell.
Advances in molecular biology in the early 1970s, including the success in creating, and transferring DNA molecules into cells, revolutionized both science and industry. The first genetically modified organisms were bacteria that made simple proteins of pharmaceutical interest, for example, insulin. As the technologies improved, other organisms including plants became amenable for improvement by rDNA technology. Table 1 provides important milestones in the development and application of rDNA technology.
Mendel’s experiments published
DNA discovered in cell
Mutation of genes by x-rays
One gene-one enzyme hypothesis
DNA is identified as the genetic material
Structure of DNA determined
Ribosomes synthesize protein
Function of mRNA proposed
Genetic code determined
Isolation of a restriction enzyme
Recombinant DNA techniques developed
Early 1970s
Isolation of a single copy gene from higher eukaryote
Rapid method of DNA sequencing developed
Plant transformation
Field testing of transformed plants
Release of engineered plants to general public in the US
Transformation of cells with rDNA produces organisms called bioengineered or genetically modified organisms (GMOs). The GMOs contain new traits from another organism. The first GMOs were Escherichia coli cells that were transformed with genes from human to produce various proteins for pharmaceutical purposes.
Isolation of DNA, restriction digestions and its separation on an agarose gel:
Preparation of dna.
For recombinant DNA procedures to work, a pure DNA sample must be obtained. The challenge is that plant and animal cells produce numerous other compounds that often act as contaminants and may inhibit cloning or sequencing of the DNA. Also, tissues and organs from the same plant, or different plants often contain different composition of metabolites, for example, proteins, lipids, and carbohydrates. These compounds must be separated from the DNA during isolation. To achieve this, scientists take advantage of the chemical and physical properties of different molecules inside the cell. For example, DNA is negatively charged, making it soluble in aqueous solution. However, the polar sugar phosphate groups of the DNA are repelled by non-polar solutions. Therefore, the final step in many DNA purifications protocols involves precipitation using alcohol. Other compounds, for example, proteins can also be easily separated from DNA by altering the concentration of salt in the extraction buffer.
Digestion of DNA with restriction endonucleases
Restriction endonucleases are a group of enzymes derived (primarily) from bacteria. Although there are several different types of restriction enzymes , those most useful for rDNA technology recognize specific short sequences in DNA and cleave the DNA at that site to produce cohesive (sticky) or blunt-ended fragments (Figure 2).

More than 500 different restriction enzymes have been identified and can be purchased commercially. Thus, one may ask the question, how often does a restriction enzyme cut within a genomic sequence? It is not possible to give an exact answer for this question. However, let us assume that there are 4 bases on any strand of DNA. This means the probability of detecting an A (adenine) at a particular location is 1/4. Now, since most restriction enzymes recognize specific sequences of 6 bases long, the probability of finding such a site is (1/4)6 = 1 site in every 4,096 base pairs (bp). Assuming you have isolated genomic DNA from maize, and you want to digest it with a restriction enzyme that cuts every 4,096 (4,100) bp, how many fragments will you obtain? To answer this question, you need to have an idea of the size of the genome of the plant you are working with. The approximate size of the maize genome is 2,500,000,000 bp. Thus, using an enzyme that cuts every 4,100 nucleotides one would expect to obtain 2,500,000,000 bp/4,100 bp, approximately 610, 000 fragments. Note that nucleotide distribution is not always random and thus frequency may be different for given DNA. Also, methylation of specific bases in genomic DNA can prevent cleavage at some site.
Separation of digested genomic DNA on agarose gel
The only physical features of nucleic acid fragments that are routinely used for their characterization are their size and nucleotide sequence. Molecular weight of DNA is most conveniently evaluated by electrophoresis in agarose gels. Agarose forms a gel by hydrogen bonding when cooled from the melted state. This gel, interwoven network of agarose chains, interferes with the movement of DNA through the gel (see Lesson on PCR and Gel Electrophoresis ). Pore size, which affects rate of movement of DNA fragments of a given size, depends on the concentration of agarose. The gel is submerged in electrolyte solution; sample is loaded into wells on one end and current is applied to facilitate movement of DNA fragments. Since DNA is negatively charged it will migrate in the electrical field. Fragments separate according to size. The distance of the migration in each time is proportional to 1/log MW. Following gel electrophoresis DNA can be visualized by staining with ethidium bromide (see Lesson on PCR and Gel Electrophoresis ) or other DNA stains.
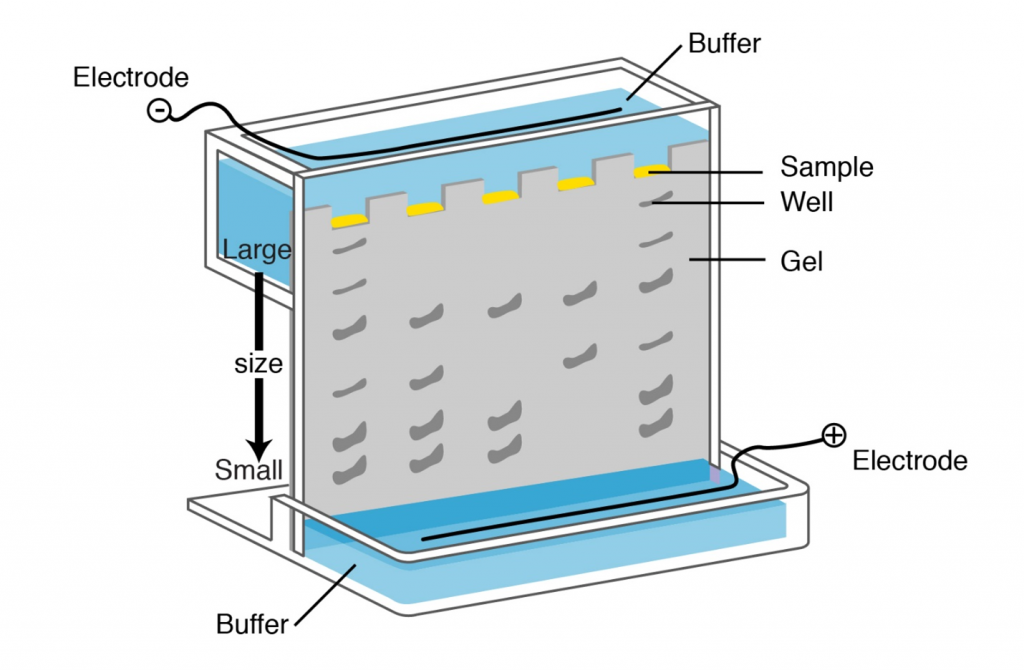
Tools for gene (DNA) cloning
Polymerase chain reaction (pcr).
Polymerase chain reaction (PCR) is a method by which millions of copies of a DNA fragment are produced in a test tube in a matter of minutes or a few hours. The basic steps in PCR reaction were discussed in the Lesson on PCR and Gel Electrophoresis.
Once the sequence of a particular gene is known, it becomes possible to use PCR to isolate that gene from any DNA sample. Recall in Lesson on Gene Transcription you learned that at the genomic level a gene is made up of regulatory sequences, coding, and non-coding sequences. Thus, if the goal is to use PCR to isolate both regulatory and non-regulatory sequences of a gene, the approach would be to use DNA as the starting material.
In cloning genes by PCR, restriction enzyme sites are added at the 5′-end of the primers to facilitate cloning of PCR fragments. A few additional nucleotides (~6 nucleotides) added at the 5′-end of the restriction sites to facilitate restriction digestion of PCR products prior to cloning in a plasmid vector. Alternatively, PCR products may be cloned directly into a T-vector without restriction digestions in E. coli (read more about Promega cloning vector systems ). After cloning into E. coli , the fragment is analyzed by sequencing and then sub-cloned into suitable vectors for expression studies.
Like all other biochemical processes, DNA synthesis by PCR is not a perfect process, and occasionally the polymerase enzyme will add an incorrect base to the growing DNA strand. In the context of DNA replication in a cell, the errors are corrected by the DNA polymerase, this is called “proofreading”. Commercially available polymerases may or may not have proofreading capability.
Another important consideration in PCR analysis is contamination.
Minor contamination of the starting material can have serious consequences. Recall that minute amounts of starting DNA can be amplified to millions of copies through PCR. If one inadvertently (or carelessly) mixed DNA from two different sources, the results will be confounding making it impossible to distinguish lines and may cost a laboratory time and money. Ensure that proper procedures are followed in preparing PCR assays. One common source of contamination in plant biology is the products from previous amplification processes. A completed PCR reaction will contain millions of copies of amplified fragments so that even a minute droplet or aerosol from a pipette tip will contain an enormous number of amplifiable molecules. It is always essential to run negative controls which will reveal the presence of contaminating DNA in your PCR assays.
Cloning vector definition and requirements
A cloning vector is a specialized DNA sequence that can enter a living cell and provide means for detection of its presence to a researcher by conferring a selectable property on the host cell (e.g., resistance to antibiotics), and possess means for self-replication. A vector must also possess easily distinguishable physical traits, such as size, or shape, to allow purification away from the host cell’s genome.
Ligase enzyme and gene cloning
Cutting and joining together of vector and DNA fragments from different origins results in rDNA. Recall that restriction endonucleases are used to cut DNA. To join DNA molecules together, an enzyme called DNA ligase is used. The enzyme DNA ligase is used to seal together restriction fragments by forming new phosphodiester bonds. The ligated vector and DNA fragment can now be transformed into a host cell for replication and expression.
The transformation of E. coli takes several steps. First, a gene of interest is inserted into a plasmid that contains a selectable marker usually encoding for resistance to an antibiotic. Second, the plasmid construct containing the gene of interest is transformed into bacterial cells by briefly exposing the mixture of ligated plasmid-DNA fragment (rDNA molecule) and bacterial cells to cold (0 o C) and heat (37-42 o C). The next step is to grow the transformed cells on selection media containing an antibiotic. Only the cells that have been transformed with the plasmid containing the gene of interest and the marker for resistance to the antibiotic will survive. In addition to using an antibiotic, plasmid vector systems that contain the lacZ gene encoding β-galactosidase allow for easier selection of positive colonies that may harbor the rDNA molecule of interest.
Types of cloning vectors
An example of a cloning vector is a plasmid (Figure 4), defined as an autonomously replicating extra chromosomal circular DNA which is faithfully passed on to progeny. Plasmids are double stranded circular DNA and range in size from about 1 kb – 200 kb. The most useful for cloning are 2 – 10 kb because smaller plasmids are easier to manipulate and usually produce higher copy numbers when grown in bacterial host cells.
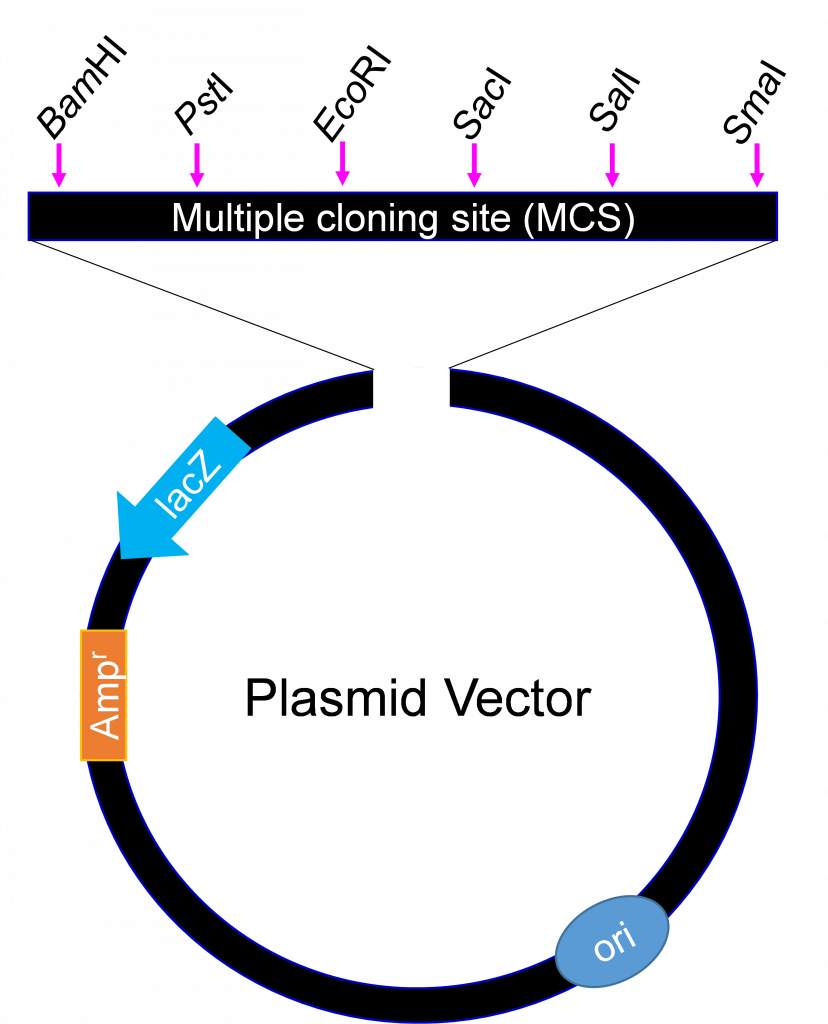
Generally, no more than 10 kb is cloned into plasmids. For cloning large DNA fragments with high efficiency, a vector called bacteriophage lambda is used. Large chromosomal DNA fragments close to 23 kb are stable when introduced into a lambda phage vector.
mRNA as starting material for gene cloning
Recall in the Lesson on Transcription you learned about synthesis of RNA from DNA through the process of transcription . Transcription is an important step in gene expression. The mRNA produced can be isolated and “copied” back to DNA by a process called reverse-transcription (Figure 5). The first step of reverse transcription mimics a strategy used by retroviruses (e.g., HIV) that have RNA genomes. As part of their gene-transmission package, retroviruses also contain an enzyme called RNA-dependent DNA Polymerases, commonly referred to as reverse transcriptase . After infecting a host cell the retrovirus uses its reverse transcriptase to copy its single stranded RNA genome into a strand of complementary DNA (cDNA). The reverse transcriptase then synthesizes the second DNA strand from the first strand to make a double stranded-DNA copy which integrates into the host genome.
If only a gene’s coding sequence is required, isolating the gene from cDNAs would be the strategy. It is important that cDNAs are synthesized from tissues expressing the gene of interest. Thus, prior knowledge of where the gene is functional is important in constructing the cDNA molecules for cloning the gene of interest.
The cDNAs produced in vitro (Figure 4) can be used for PCR analysis, similar to chromosomal DNA. The combination of reverse-transcription and PCR (RT-PCR) is a valuable tool in gene cloning and quantification of mRNA.
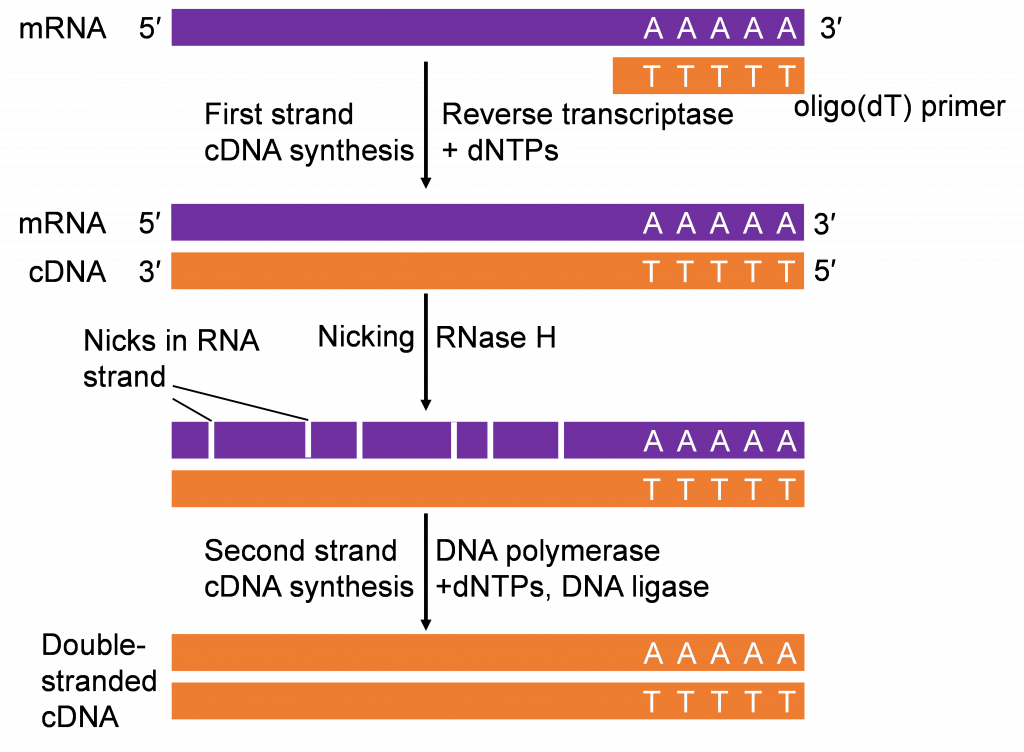
Recombinant DNA technology has contributed significantly to development of agricultural biotechnology. Transformation of cells with rDNA produces organisms called bioengineered or genetically modified organisms. The tools for plant rDNA technology include, vectors, restriction enzyme, ligation enzymes, bacterial hosts, methods to isolate and multiply nucleic acids, methods to quantify nucleic acids, Agrobacterium as a vector to insert foreign DNA into plants.
Below is a hypothetical DNA fragment containing restriction sites for EcoRI and BamHI.
- How many fragments will be produced after cutting the DNA with BamHI? What size will these fragments be in bp?
- How many fragments will be produced after cutting the DNA with both BamHI and EcoRI at the same time? What size will these fragments be in bp?
- How many fragments will be produced after cutting the DNA with EcoRI? What size will these fragments be in bp?
- You need to clone an EcoRI fragment in the EcoRI site of an expression plasmid vector in E coli. The fragment is 1,809 bp long. There is a BamHI site in the fragment at 1,204 bp. In the plasmid vector, there is a single BamHI site at the promoter region for expression of the gene in E. coli. The BamHI site is 100 bp upstream of EcoRI cloning site.
Following cloning the EcoRI fragment you will get two classes of clones, only one of which will produce the protein. Describe the approach you will take to identify the correct class of clones for expression of the gene in E. coli.
Browse Course Material
Course info, instructors.
- Prof. Barbara Imperiali
- Prof. Adam Martin
- Dr. Diviya Ray
Departments
As taught in.
- Functional Genomics
- Biochemistry
- Cell Biology
- Microbiology
- Molecular Biology
Learning Resource Types
Introductory biology, lecture 16: recombinant dna, cloning, & editing, description.
In today’s lecture, the focus shifts from pure genetics to molecular genetics, beginning with cloning, followed by polymerase chain reaction (PCR), and finally genome editing.
Instructor: Adam Martin
- Download video
- Download transcript

You are leaving MIT OpenCourseWare
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
DNA recombination articles from across Nature Portfolio
DNA recombination is the exchange of DNA strands to produce new nucleotide sequence arrangements. Recombination occurs typically, though not exclusively, between regions of similar sequence by breaking and rejoining DNA segments, and is essential for generating genetic diversity and for maintaining genome integrity.

Latest Research and Reviews
Seeding the meiotic DNA break machinery and initiating recombination on chromosome axes
Meiotic cells deliberately break their DNA to allow chromosomes to swap genetic material. Here, authors reveal genetically separable pathways controlling the seeding and growth of chromosome-bound protein condensates responsible for DNA breaks.
- Ihsan Dereli
- Vladyslav Telychko
- Attila Tóth
Dbf4-dependent kinase promotes cell cycle controlled resection of DNA double-strand breaks and repair by homologous recombination
The repair of DNA double strand breaks is strictly controlled during the cell cycle by the CDK kinase. Here the authors identify the DDK kinase as a second major regulator for this cell cycle regulation and elucidate its functional targets.
- Lorenzo Galanti
- Martina Peritore
- Boris Pfander
Engineering intelligent chassis cells via recombinase-based MEMORY circuits
The unification of decision-making, communication, and memory would enable the programming of intelligent biotic systems. Here, the authors achieve this goal by engineering E. coli chassis cells with an array of inducible recombinases that mediate diverse genetic programs.
- Brian D. Huang
- Corey J. Wilson
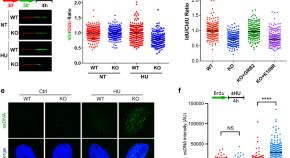
GRB2 stabilizes RAD51 at reversed replication forks suppressing genomic instability and innate immunity against cancer
GRB2 is known for its role in Receptor Tyrosine Kinase and RAS signaling. Here the authors unveil a GRB2 function and mechanism for DNA replication fork protection. GRB2 alleviates oncogenic replication stress, and in doing so, averts cancer immune destruction by inhibiting cGAS/STING and pro-inflammatory cytokine production.
- Shengfeng Xu
- Zamal Ahmed
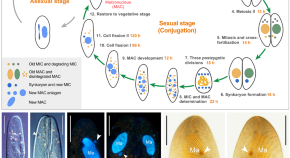
Application of RNA interference and protein localization to investigate housekeeping and developmentally regulated genes in the emerging model protozoan Paramecium caudatum
Methods of gene silencing by RNAi and protein localization by microinjection of fusion constructs have been established in the emerging model protozoan Paramecium caudatum . These methods lay the groundwork for future studies of gene function.
- Therese Solberg
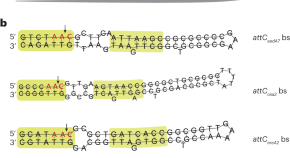
Integron cassettes integrate into bacterial genomes via widespread non-classical attG sites
Computational analyses and molecular genetics reveal that integron cassettes integrate into bacterial genomes at widespread non-classical attG sites, enabling acquisition of genetic material and disruption of existing genes with potential consequences for bacterial evolution.
- Céline Loot
- Gael A. Millot
- Didier Mazel
News and Comment

Competition between sites of meiotic recombination in snakes
A study in Science reports that corn snakes use both PRDM9 and promoter-like features to direct meiotic recombination, indicating that these are not mutually exclusive.
- Kirsty Minton
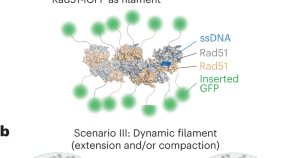
Visualizing the repair of broken DNA in living cells reveals a new mechanism for DNA homology search
We engineered a tagged version of the yeast Rad51 recombinase and used this tool to monitor DNA double-strand break repair in living cells. We could observe how a broken DNA fragment can scout the nucleus to identify a similar sequence and use it as a template for repair.
Loss-of-function variants in human C12orf40 cause male infertility by blocking meiotic progression
- Chaofeng Tu
- Yue-Qiu Tan
DNA scaffolds encoding messages
The Rad52 SSAP superfamily and new insight into homologous recombination
Recent structures of DNA-bound bacterial and phage recombinases provide insights into homologous recombination and suggest relation to the eukaryotic Rad52 and identification of a Rad52 single strand annealing protein (SSAP) superfamily.
- Ali Al-Fatlawi
- Michael Schroeder
- A. Francis Stewart
Dynamic control of the meiotic recombination landscape
Steiner highlights a study by Protacio et al., which has identified molecular mechanisms underlying the plasticity of meiotic recombination in Schizosaccharomyces pombe .
- Walter Steiner
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Introduction to Genomics
- Educational Resources
- Policy Issues in Genomics
- The Human Genome Project
- Funding Opportunities
- Funded Programs & Projects
- Division and Program Directors
- Scientific Program Analysts
- Contact by Research Area
- News & Events
- Research Areas
- Research investigators
- Research Projects
- Clinical Research
- Data Tools & Resources
- Genomics & Medicine
- Family Health History
- For Patients & Families
- For Health Professionals
- Jobs at NHGRI
- Training at NHGRI
- Funding for Research Training
- Professional Development Programs
- NHGRI Culture
- Social Media
- Broadcast Media
- Image Gallery
- Press Resources
- Organization
- NHGRI Director
- Mission & Vision
- Policies & Guidance
- Institute Advisors
- Strategic Vision
- Leadership Initiatives
- Diversity, Equity, and Inclusion
- Partner with NHGRI
- Staff Search
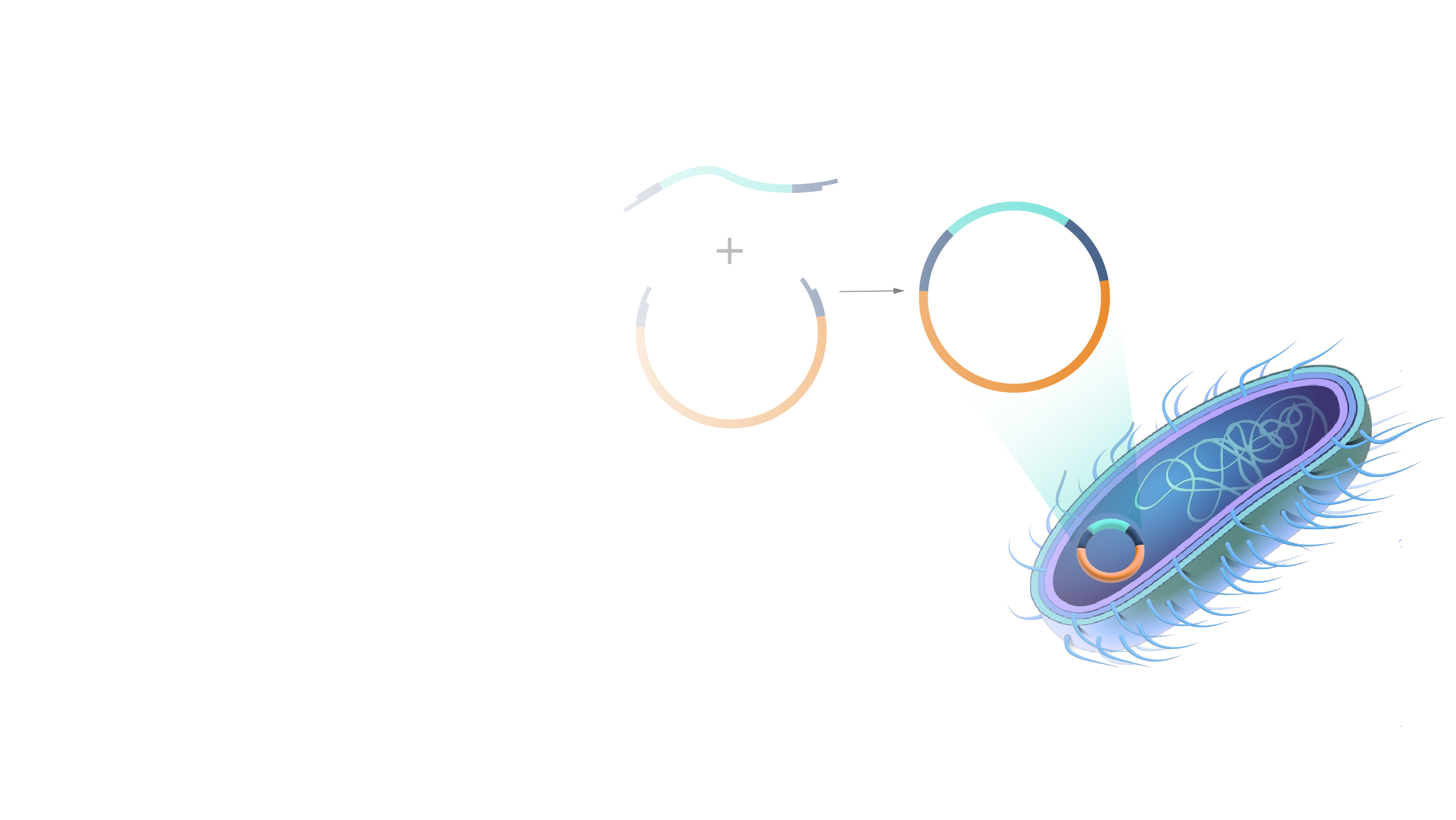
Recombinant DNA Technology
Recombinant DNA technology involves using enzymes and various laboratory techniques to manipulate and isolate DNA segments of interest. This method can be used to combine (or splice) DNA from different species or to create genes with new functions. The resulting copies are often referred to as recombinant DNA. Such work typically involves propagating the recombinant DNA in a bacterial or yeast cell, whose cellular machinery copies the engineered DNA along with its own.
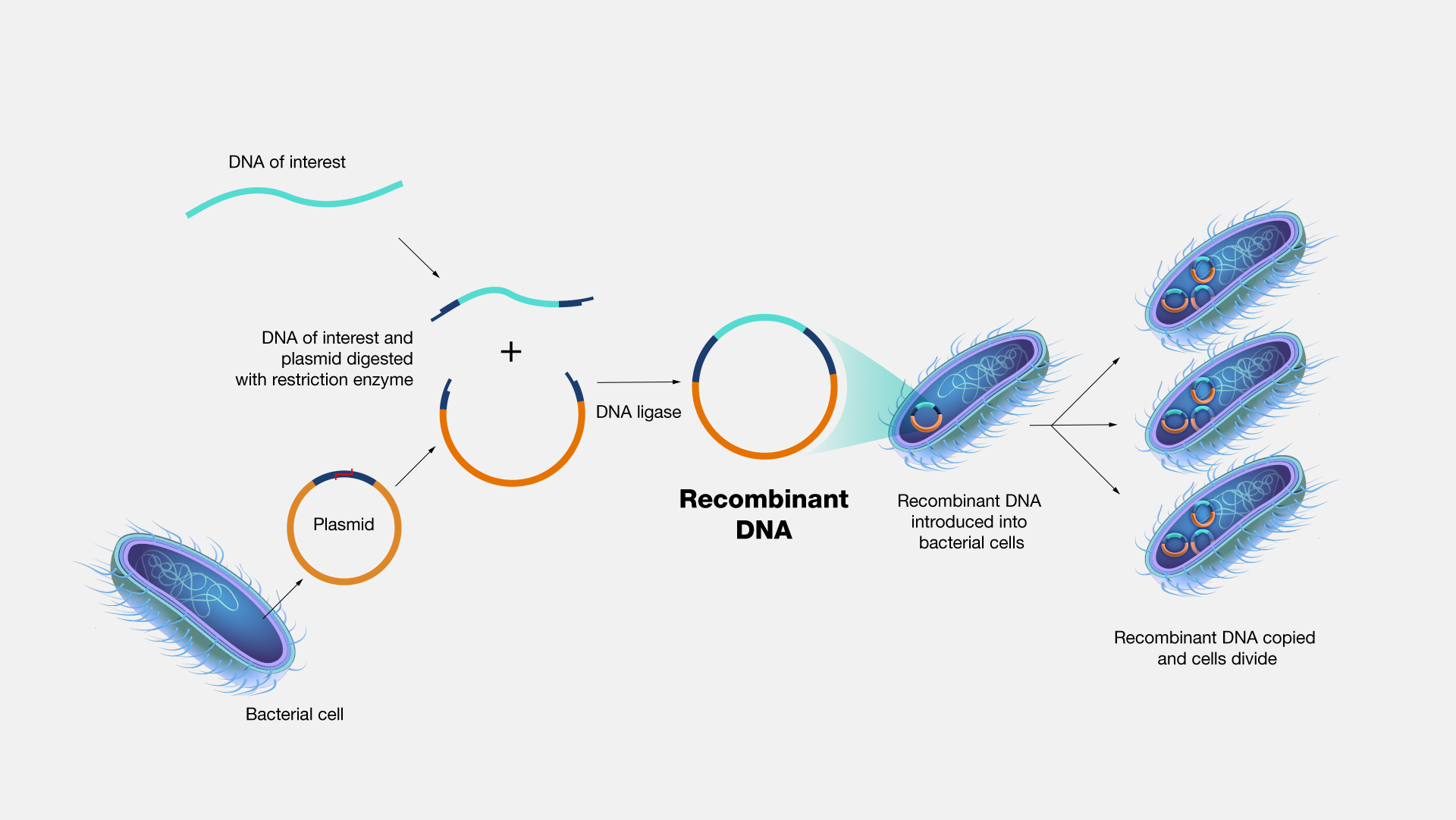
Recombinant DNA Technology. Recombinant DNA technology is an extremely important research tool in biology. It allows scientists to manipulate DNA fragments in order to study them in the lab. It involves using a variety of laboratory methods to put a piece of DNA into a bacterial or yeast cell. Once in, the bacteria or yeast will copy the DNA along with its own. Recombinant DNA technology has been successfully applied to make important proteins used in the treatment of human diseases, such as insulin and growth hormone.
Scientific Liaison to the Director for Extramural Activities
Office of the Director
Recombinant DNA Technology
Jan 01, 2020
560 likes | 1.62k Views
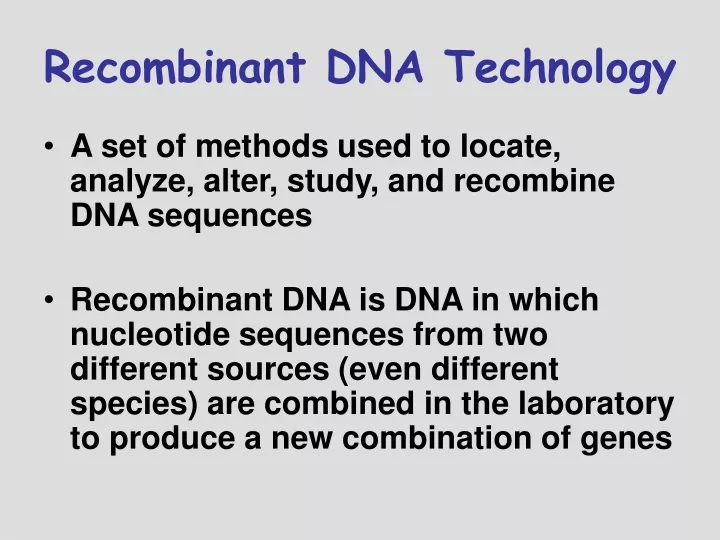
Recombinant DNA Technology. A set of methods used to locate, analyze, alter, study, and recombine DNA sequences Recombinant DNA is DNA in which nucleotide sequences from two different sources (even different species) are combined in the laboratory to produce a new combination of genes.
Share Presentation
- dna sequence
- recombinant dna
- dna sequence accomplishes
- staggered cuts sticky ends

Presentation Transcript
Recombinant DNA Technology • A set of methods used to locate, analyze, alter, study, and recombine DNA sequences • Recombinant DNA is DNA in which nucleotide sequences from two different sources (even different species) are combined in the laboratory to produce a new combination of genes
Recombinant DNA Technology • It is used to: • Probe the structure and function of genes • If I delete this gene, what happens to the mouse? • Address questions in many areas of biology • How are Bos taurus and Bos indicus related? • Create commercial products • How can I get insulin production from bacteria? • Diagnose and treat diseases • Does this child have cystic fibrosis?
Recombinant DNA Technology • Using Mendelian and other genetic analyses we have talked about, everything we have deduced about genes and DNA sequence has been indirect • With recombinant DNA technology we can isolate genes and DNA sequence, study them directly andstore it in a convenient manner that facilitates future applications • Cloning the DNA sequence accomplishes all of these
Review of steps involved in cloning • http://highered.mcgraw-hill.com/sites/0072556781/student_view0/chapter14/animation_quiz_1.html
Recombinant DNA Technology • Restriction enzymes were the key breakthrough in recombinant DNA technology and cloning • Restriction enzymes are used to cleave DNA at specific sequences • Cuts DNA at specific sequences into different sized molecules • Cuts a single phosphodiester bond on each strand
Recombinant DNA Technology • Recognition sequence • Usually four to six bases • Palindromes – read the same on both strands • Cut DNA reciprocally at specific recognition sites
Recombinant DNA Technology • Most restriction enzymes make offset cuts • Restriction sites are not opposite each other • Results in staggered cuts (sticky ends) • Ability for hydrogen bonding to a complementary sequence
- More by User

Recombinant DNA Technology. rDNA Technology. Restriction Enzymes and DNA Ligase Plasmid Cloning Vectors Transformation of Bacteria Blotting Techniques. Restriction Enzymes. Most significant advancement permitting rDNA manipulation Differ from other nucleases
1.87k views • 57 slides

Recombinant DNA Technology. Some basic terminology…. Recombinant DNA (rDNA)- contains DNA from 2 sources; DNA is inserted into that of another organism Vector- DNA piece that can be modified to accommodate a piece of foreign DNA
1.21k views • 10 slides

Recombinant DNA Technology………..
Recombinant DNA Technology………. BTEC3301. DNA Libraries. How do you identify the gene of interest and clone only the DNA sequence you are interested? Read Pg 63. DNA Libraries. Library screening of gene of interest
593 views • 17 slides

Recombinant DNA Technology. Prof. Elena A. Carrasquillo Chapter 4 Molecular Biotechnology Lecture 4. Cloning in Plasmid Vector In bacteriophage vector Screening the library DNA probe Antibody probe. Major Steps in building a DNA Library.
1.79k views • 43 slides

Harini Chandra. Recombinant DNA Technology.
505 views • 26 slides

Recombinant DNA Technology. Recombinant DNA Technology. T echnique that allows DNA to be combined from different sources DNA code universal Foundation of genetic engineering. Review. What are enzymes? What function do they serve? What makes up the DNA sequence?
1.27k views • 45 slides

Recombinant DNA Technology. Needles in Haystacks. How to find one gene in large genome? A gene might be 1/1,000,000 of the genome. Three basic approaches: 1. Polymerase chain reaction ( PCR ). Make many copies of a specific region of the DNA.
526 views • 33 slides
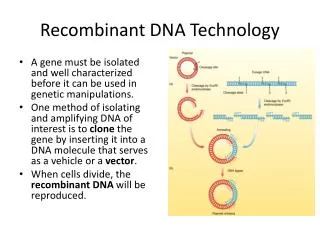
Recombinant DNA Technology. A gene must be isolated and well characterized before it can be used in genetic manipulations. One method of isolating and amplifying DNA of interest is to clone the gene by inserting it into a DNA molecule that serves as a vehicle or a vector .
4.66k views • 31 slides

Recombinant DNA Technology. Review. What are enzymes? What function do they serve? What makes up the DNA sequence? How is the genetic code determined in an organism? . Recombinant DNA Technology. http://www.youtube.com/watch?v=x2jUMG2E-ic. Discovery of restriction enzymes.
792 views • 41 slides

Recombinant DNA Technology. What is different between these 2 sequences? GGAATTCCTAGCAAT CCTTAAGGATCGTTA CTACGTGAGGAATTC GATGCACTCCTTAAG. Only order of bases!
324 views • 9 slides

DNA Recombinant Technology
DNA Recombinant Technology. What and Why?. What?: A gene of interest is inserted into another organism, enabling it to be cloned, and thus studied more effectively
468 views • 26 slides

Recombinant DNA technology
Recombinant DNA technology. BIO3B. Revision. Draw a label to identify: The template strand The coding strand A nucleotide A hydrogen bond A codon An amino acid Transcription Translation Describe the roles of Ribosomes tRNA RNA polymerase. Revision. The coding strand.
406 views • 17 slides

DNA Recombinant Technology. DNA recombinant. Genetic Engineering. The manipulation of an organism endowment by introducing or eliminating specific gene A gene of interest is inserted into another organism, enabling it to be cloned, and thus studied more effectively
777 views • 50 slides

RECOMBINANT DNA TECHNOLOGY
RECOMBINANT DNA TECHNOLOGY. PART 2. Topics. Genomic libraries Id a specific clone or sequence within a library Transgenic plants Transgenic animals Timely topics. Genomic library contains all of the organism’s genome. Probe design. Bacterial clones and finding desired clone.
487 views • 29 slides

Recombinant DNA Technology. r-DNA techniques. Isolation of Cellular DNA and RNA Plasmids Ability to disrupt cells and recover intact, pure DNA (or RNA) Reverse Transcriptase Restriction enzymes Sequence specific DNA cutting reactions Transformation techniques CaCl2 method Electroporation
492 views • 15 slides

Recombinant DNA Technology. BTEC3301. Recombinant DNA Technology.
506 views • 39 slides

BIOCHEMISTRY
665 views • 48 slides

Recombinant DNA Technology. Reference Books. Hortons’ Principles of Biochemistry 4th edition Molecular Biology , Robert Weaver 2nd edition Harpers’ Biochemistry 26th edition Styers’ Biochemistry. Objectives:
1.36k views • 119 slides
DNA recombinant technology
DNA recombinant technology. Definition of recombinant DNA technology( 基因重組 ). A series of procedures used to recombine DNA segments.. Under certain conditions, a recombinant DNA molecule can enter a cell and replicate. History of recombinant DNA technology.
429 views • 20 slides

Recombinant DNA Technology. replication. replication. transcription. transcription. translation. translation. Gene expression in vivo. DNA from different source. DNA Recomnbinant. DNA Recombination. Development of clone technique. 1. Restriction Enzymes and DNA Ligase Are Key
1.15k views • 93 slides

401 views • 33 slides
Recombinant DNA technology DNA has now become the easiest of the cellular macromolecules to be analyzed
601 views • 42 slides

- Recombinant DNA Technology
- Popular Categories
Powerpoint Templates
Icon Bundle
Kpi Dashboard
Professional
Business Plans
Swot Analysis
Gantt Chart
Business Proposal
Marketing Plan
Project Management
Business Case
Business Model
Cyber Security
Business PPT
Digital Marketing
Digital Transformation
Human Resources
Product Management
Artificial Intelligence
Company Profile
Acknowledgement PPT
PPT Presentation
Reports Brochures
One Page Pitch
Interview PPT
All Categories
Powerpoint Templates and Google slides for Recombinant DNA Technology
Save your time and attract your audience with our fully editable ppt templates and slides..
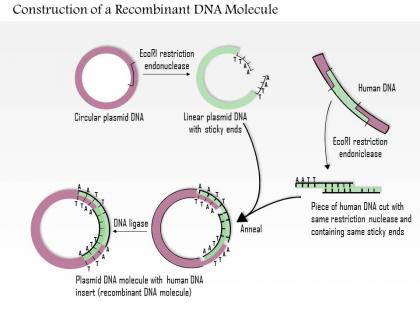
We are proud to present our 0614 construction of a recombinant dna molecule medical images for powerpoint. Construction of DNA molecules is used in this Medical Power Point template. Show recombinant of these cells in your presentation and get full remarks from your viewers.

Presenting our Recombinant Dna Technology Ppt Powerpoint Presentation Slides Examples Cpb PowerPoint template design. This PowerPoint slide showcases five stages. It is useful to share insightful information on Recombinant DNA Technology This PPT slide can be easily accessed in standard screen and widescreen aspect ratios. It is also available in various formats like PDF, PNG, and JPG. Not only this, the PowerPoint slideshow is completely editable and you can effortlessly modify the font size, font type, and shapes according to your wish. Our PPT layout is compatible with Google Slides as well, so download and edit it as per your knowledge.

Presenting this set of slides with name DNA Recombinant Technology For Medical Science. The topics discussed in these slide is DNA Recombinant Technology For Medical Science. This is a completely editable PowerPoint presentation and is available for immediate download. Download now and impress your audience.
Presenting our Cell Transformation Successful Recombinant Dna Ppt Powerpoint Presentation Summary Brochure Cpb PowerPoint template design. This PowerPoint slide showcases four stages. It is useful to share insightful information on Cell Transformation Successful Recombinant Dna This PPT slide can be easily accessed in standard screen and widescreen aspect ratios. It is also available in various formats like PDF, PNG, and JPG. Not only this, the PowerPoint slideshow is completely editable and you can effortlessly modify the font size, font type, and shapes according to your wish. Our PPT layout is compatible with Google Slides as well, so download and edit it as per your knowledge.


IMAGES
VIDEO
COMMENTS
rDNA Technology: DNA Manipulation for Research & Applications. May 3, 2015 •. 523 likes • 267,576 views. AI-enhanced title and description. nasira jaffry. This document provides an overview of recombinant DNA technology. It discusses the key discoveries that led to the development of this technology, such as Watson and Crick's discovery of ...
During download, if you can't get a presentation, the file might be deleted by the publisher. E N D . ... RECOMBINANT DNA TECHNOLOGY. PART 2. Topics. Genomic libraries Id a specific clone or sequence within a library Transgenic plants Transgenic animals Timely topics. Genomic library contains all of the organism's genome.
1.03.7 Conclusions. Recombinant DNA technology has offered a complete solution for cloning and expressing a gene of interest. Bioprocessing strategies for fermentation and downstream processing have been well developed during the past few decades to the point where mass-producing an enzyme of interest becomes routine.
The complete process of recombinant DNA technology includes multiple steps, maintained in a specific sequence to generate the desired product. Step-1. Isolation of Genetic Material. The first and the initial step in Recombinant DNA technology is to isolate the desired DNA in its pure form i.e. free from other macromolecules. Step-2.
To print or download this file, click the link below: Microbiology Chapter 8.pptx — application/vnd.openxmlformats-officedocument.presentationml.presentation, 5.99 ...
The possibility for recombinant DNA technology emerged with the discovery of restriction enzymes in 1968 by Swiss microbiologist Werner Arber.The following year American microbiologist Hamilton O. Smith purified so-called type II restriction enzymes, which were found to be essential to genetic engineering for their ability to cleave at a specific site within the DNA (as opposed to type I ...
Definition and background. Recombinant rDNA technology involves procedures for analyzing or combining DNA fragments from one or several organisms (Figure 1) including the introduction of the rDNA molecule into a cell for its replication, or integration into the genome of the target cell. Figure 1. Recombinant DNA is made from combining DNA from ...
During this unit, you will learn the steps involved in a basic cloning strategy. You will determine which restriction enzyme to use to create a desired piece of recombinant DNA, given specific DNA sequences. You will also learn the function of DNA ligase, understand how vectors are used, and learn how to construct a recombinant genomic DNA library.
Description. In today's lecture, the focus shifts from pure genetics to molecular genetics, beginning with cloning, followed by polymerase chain reaction (PCR), and finally genome editing.
Recombinant DNA is the method of joining two or more DNA molecules to create a hybrid. The technology is made possible by two types of enzymes, restriction endonucleases and ligase. A restriction endonuclease recognizes a specific sequence of DNA and cuts within, or close to, that sequence. By chance, a restriction enzyme's recognition sequence ...
DNA recombination is the exchange of DNA strands to produce new nucleotide sequence arrangements. Recombination occurs typically, though not exclusively, between regions of similar sequence by ...
Recombinant DNA technology was first applied to protein production in mammalian cells in the early 1980s. This approach was facilitated by the development of transfection methods for the efficient delivery of plasmid DNA into cultivated mammalian cells. Chinese hamster ovary (CHO) cells were the first mammalian host to be used for gene transfer ...
This ppt tell us about the key tool and processes used in recombinant DNA technology. Education. 1 of 32. Download Now. Download to read offline. Recombinant DNA Technology - Download as a PDF or view online for free.
2. Recombinant DNA Technology. Recombinant DNA technology comprises altering genetic material outside an organism to obtain enhanced and desired characteristics in living organisms or as their products. This technology involves the insertion of DNA fragments from a variety of sources, having a desirable gene sequence via appropriate vector .
Definition. Recombinant DNA technology involves using enzymes and various laboratory techniques to manipulate and isolate DNA segments of interest. This method can be used to combine (or splice) DNA from different species or to create genes with new functions. The resulting copies are often referred to as recombinant DNA.
D. Amplifying the recombinant DNA To recover large amounts of the recombinant DNA molecule, it must be amplified. This is accomplished by transforming the recombinant DNA into a bacterial host strain. (The cells are treated with CaCl 2 à DNA is added à Cells are heat shocked at 42 C à DNA goes into cell by a somewhat unknown mechanism.)
Presentation Transcript. Manupulation of DNA - isolation and cloning Stages: 1. Generation of DNA fragments • By splicing by restriction endonucleases [RE]. • Splicing DNA fragment with sticky or blunt ends. 2. Separation of DNA fragments - by agarose gel electrophoresis/PAGE- Selection of desired piece of DNA 3.
Recombinant DNA (rDNA) technology involves combining DNA molecules from different sources into a single recombinant DNA molecule. This is done using restriction enzymes to cut the DNA at specific sites and DNA ligase to join the fragments. The resulting recombinant DNA can be inserted into a host cell that will replicate it, allowing mass ...
Recombinant DNA Technology. A set of methods used to locate, analyze, alter, study, and recombine DNA sequences Recombinant DNA is DNA in which nucleotide sequences from two different sources (even different species) are combined in the laboratory to produce a new combination of genes. Download Presentation. dna.
The topics discussed in these slide is DNA Recombinant Technology For Medical Science. This is a completely editable PowerPoint presentation and is available for immediate download. Download now and impress your audience. Slide 1 of 2. Cell transformation successful recombinant dna ppt powerpoint presentation summary brochure cpb.